Base-pairing rule
n., plural: base-pairing rules
[beɪsˈpɛərɪŋ ɹuːl]
Definition: rule that applies to the pairing of the nitrogenous bases in the nucleic acid
Table of Contents
Base-pairing Rules Definition
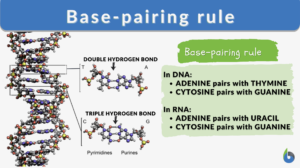
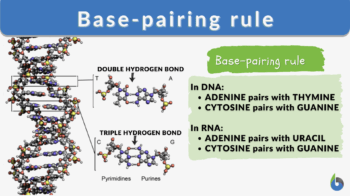
The base-pairing rules are rules that apply during the pairing between one purine and one pyrimidine via tight hydrogen bonds (H-bonds).
- These rules apply to both types of double-stranded nucleic acids, particularly, double-stranded DNA as well as double-stranded RNA (with an exception of A = T in DNA and A = U in RNA).
- The DNA base-pairing rules are strict in the sense that no nucleotide pairs with any other nucleotide except for its designated partner. The designated partners form complementary base pairs with each other as follows:
- Another important thing to learn here is that these bonds are typical for the nucleotides chosen. This means that according to DNA base pairing rules, Purine A bonds with Pyrimidine T via a double hydrogen bond. Purine G bonds with Pyrimidine C via a triple hydrogen bond.
- Because of the base-pairing in DNA, the percentage of A is equal to T and the percentage of G is equal to C.
The above rules are for DNA base pairing. In RNA pairing, it is slightly different.
- In RNA, cytosine (C) pairs with guanine (G) (the same way as in DNA)
- In RNA, adenine pairs with uracil (U) in contrast to pairing with thymine (T) in DNA.
(For an overview of the basic concepts of DNA, definition, composition, and difference from another nucleic acid, RNA, jump to: DNA Basics)
You’re probably wondering, why not adenine and cytosine? … or guanine and thymine? … How about adenine and guanine? … or thymine and cytosine? The answer to that has to do with the available space. Pairing between purines (or two pyrimidines) would take up too much space; such pairings will not fit in the two strands and so, this prevents them to pair up.
Adenine pairing up with thymine and cytosine pairing up with guanine are the only possible pairings that can form hydrogen bonds in that space. (Walsh, 2019) Also, remember that A=T forms via two hydrogen bonds and G≡C, via three hydrogen bonds. And so, swapping of pairs is unlikely.
Watch this vid about base pairing:
The base-pairing rule is a set of rules for the regulated form of base pairing between one purine and one pyrimidine via tight hydrogen bonds in nucleic acids like DNA or RNA.
DNA Basics
DNA, which stands for deoxyribonucleic acid, is a genetic material that is made up of four nucleotides i.e., dATP, dGTP, dCTP, and TTP (deoxyadenosine triphosphate, deoxyguanosine triphosphate, deoxycytidine triphosphate, and thymidine triphosphate, respectively).
All nucleotides are made up of three basic components as follows:
- Nitrogenous bases (four DNA bases namely adenine, thymine, cytosine, and guanine)
- Sugar (deoxyribose sugar moiety)
- Phosphate groups
Once these “nitrogenous bases in DNA” combine with the sugar moiety, they are now referred to as “nucleosides”. Only after the addition of phosphate groups to nucleosides that “nucleotides” form.
As we know that DNA is a double-stranded structure, the nucleotides on one strand form the nitrogenous base pairs with the nucleotides on the other strand. This DNA pairing ensures that the two DNA strands are bound together by tight hydrogen bonds. As commonly known, these bonding reactions are very specific and follow universal rules. These rules are called base-pairing rules.
DNA and RNA are examples of nucleic acids. However, DNA has deoxyribose sugar and RNA possesses ribose sugar. Thus, the nucleotides that form the RNA are ATP, GTP, CTP, and UTP. And as already mentioned in the previous section, RNA base pairs are also quite different from DNA base pairs in such a way that instead of thymine, adenine pairs with uracil.
Given the information above, you could answer by now common questions like what is the base pair rule, how are RNA nucleotides different from DNA nucleotides, and what is the complementary base pairing rule. Let’s learn more about the topic by understanding the historical account of base-pairing rules (DNA Complementarity) and the application of base-pairing rules, such as in the Human Genome Project.
DNA Complementarity
The discussion about “DNA complementarity” is an interesting topic since we have referred to the term ‘complementary DNA strands’ too many times here.
The principle of complementarity for different nucleic acids like DNA and RNA can be elucidated as a ‘lock & key principle’. This means that the two strands of the nucleic acids need to have an ascribed structure that fits into one another like a lock and its very specific key. This ascribed structure is attained via distinct and specific hydrogen bonding interactions between nucleotides.
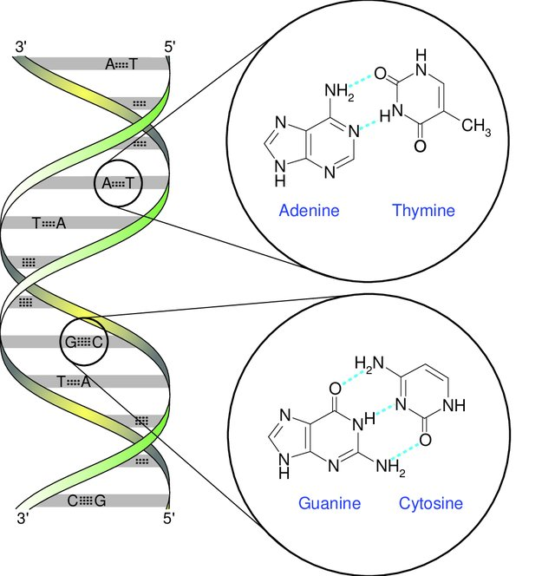
The double H-bonding between A = T and the triple hydrogen bonding between G ≡ C pave way for the two DNA strands to be exact complements of each other (in an antiparallel orientation). It is this complementarity (DNA pairing) only that ensures the formation of the double helix structure of DNA.
The double helix structure was an original idea proposed by a female scientist “Rosalind Franklin” who deciphered it via her X-ray crystallography experiments.
Later, Watson and Crick described it in detail. So, sometimes this DNA pairing is also referred to as “Watson-Crick Base Pairing”. The two complementary DNA strands have opposite directions and are thereby sometimes also referred to as “antiparallel” in their nature.

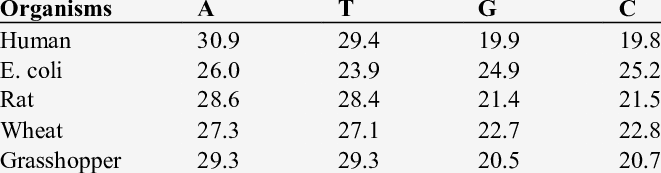
Relative Proportions (%) of Bases in DNA
It is intriguing to learn about the relative amounts of different nucleotides in the DNA. Since the DNA strands are complementary, a rule called “Chargaff’s Rule” states the amount of adenine in the DNA of a living organism is equal to the amount of thymine in the DNA. It also states that the amount of cytosine in the DNA of a living being is equal to the amount of guanine in the DNA.

These rules are based on the fact that if you read one strand of the dsDNA in a 5′ to 3′ direction, you can decode the other strand very easily. It is only the (C+G): (A+T) ratio that varies from one organism to another.
The Human Genome Project
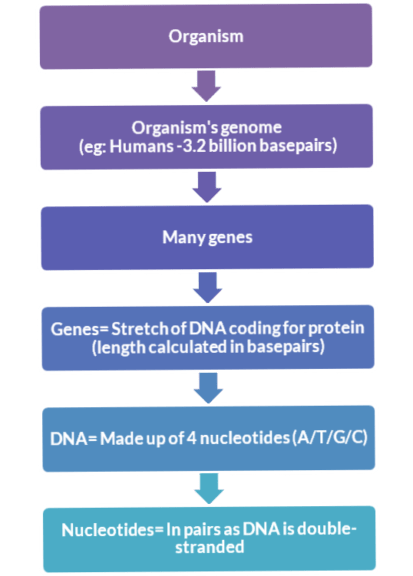
When the Human Genome Project (HGP) kicked off in 1990, no one had the minutest idea that by the time the project completes; we’ll end up with 3.2 billion DNA base pairs. Even to date, we consider the decoding of the human genome as one of the greatest and the finest endeavors that have been undertaken by the scientific community.
As we all know that every character, trait, appearance, and behavior, no matter what phenotype you bring to the table of discussion, each one has a genotypic coding in the form of DNA nucleotides base pairs in all the organisms. To make it simple, look at the hierarchy of the following flowchart:
Interesting Fact
Do you know that as large as 99.9% of the human genome is common to all human beings?
Yes, you heard it right! You share 99.9% similarity with every human around you whether it be your mom, brother, neighbor, fruit vendor, a person from a different city, or a traveler from a different continent!! You must be astonished because we think the differences are too many at the genetic level, but they aren’t. The remaining 0.1% comprises all the variations with which you differentiate yourself from your mom, neighbor, stranger, etc. These 0.1% varying regions are scientifically termed “genomic variants”. With 3.2 billion base pairs and thereby nearly 6 billion nucleotide bases or letters (HUGE number) in the entire genome, each one of us has a massive count of these genomic variants. Nearly 4-5 million genomic variants are present in each one of us…

Answer the quiz below to check what you have learned so far about base-pairing rules.
Reference
- National Human Genome Research Institute- https://www.genome.gov/Health/Genomics-and-Medicine/Polygenic-risk-scores#one
- Hood, L; Galas, D (Jan 23, 2003). “The digital code of DNA”. Nature. 421 (6921): 444–8.
- Human Genome Project- https://www.genome.gov/human-genome-project#:~:text=The%20Human%20Genome%20Project%20was,than%20the%20original%20estimated%20budget.
- Elson D, Chargaff E (1952). “On the deoxyribonucleic acid content of sea urchin gametes”. Experientia. 8 (4): 143–145. doi:10.1007/BF02170221
- Chargaff E, Lipshitz R, Green C (1952). “Composition of the deoxypentose nucleic acids of four genera of sea-urchin”. J Biol Chem. 195 (1): 155–160. doi:10.1016/S0021-9258(19)50884-5
- Walsh, E. (2019). What Is the Complementary Base Pairing Rule? (2019). Sciencing. https://sciencing.com/complementary-base-pairing-rule-8728565.html
©BiologyOnline.com. Content provided and moderated by Biology Online Editors.