Centromere
n., plural: centromeres
[ˈsɛntɹəˌmiɚ/]
Definition: the region in a chromosome where kinetochore is located
Table of Contents
Centromere Definition
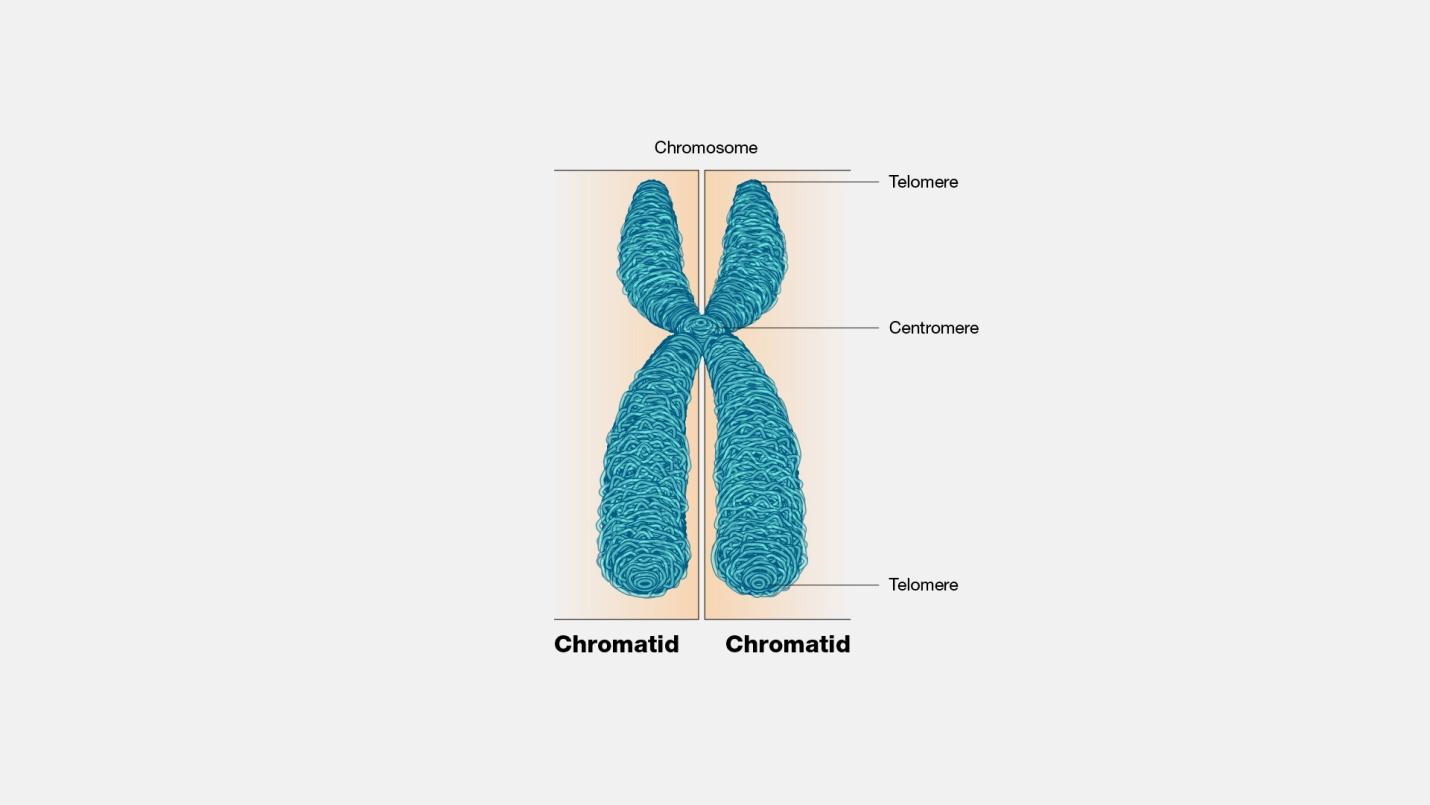
Centromere is defined as the point of attachment for the sister chromatids generated after DNA replication. When a chromosome replicates and forms two identical copies during DNA replication, the copies are referred to as “sister chromatids”. Sometimes, these sister chromatids are also called “half chromosomes”. And the point where these half-chromosomes join to form a single chromosome is called the centromere region.
Watch this vid about centromere:
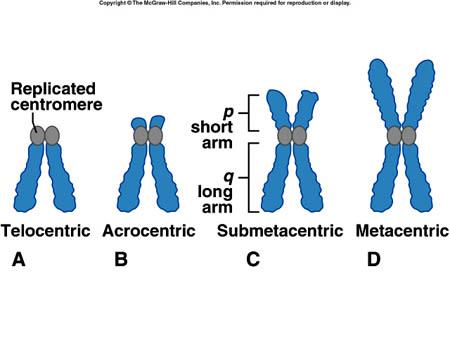
A centromere is the dense and constricted region of a chromosome. It is where the sister chromatids attach after DNA replication and kinetochore assembly (for spindle fiber attachment) take place before mitosis. It contains highly specialized repetitive DNA sequences (e.g. satellite DNA) of a chromosome linking sister chromatids forming a dyad. Most eukaryotes have DNA sequences in their centromere that are packaged into heterochromatin. When the centromere is not functioning properly, the chromatids cannot align and separate properly, thus, resulting in the wrong number of chromosomes in the daughter cells, and conditions such as Down syndrome. The position of the centromere in the chromosome results in the characterization of the chromosomal arms. The chromosomal arm that is shorter is called p whereas the longer arm is called q. Based on the position of the centromere, the chromosomes may be classified as metacentric, submetacentric, acrocentric, telocentric, subtelocentric, and holocentric.
Etymology: Latin centrum, Greek kentron + meros (“part”).
Related form(s): centromeric (adjective, of, or pertaining to, the centromere)
See also: chromatid, chromosome, kinetochore
Position
While discerning the karyotype of an individual or species, the centromere plays a very important role. Chromosomal karyotypes encompass the general shape, size, and number (i.e. appearance) of the different chromosomes (not the same chromosome) in an individual or species.
Chromosomal karyotype in human beings (human cells) is defined by the “centromere position”. This position makes two chromosome arms; one short arm (p; name derived from the French word “petite” meaning ‘small’) and one long arm (q). Based on this idea of centromere positioning, chromosomes are classified into five types for ease of study. They are metacentric, submetacentric, acrocentric, telocentric, and subtelocentric.
Metacentric
Metacentric chromosomes are characterized by their “X-shaped structure” due to the positioning of the centromere nearly at the midpoint from both ends. This results in equal-sized arms (p and q).
Centromere position: Medial sensu strict or medial region
Arm length ratio: 1.0-1.6 or 1.7
Denoted by (sign): M or mSubmetacentric
Submetacentric chromosomes are characterized by their “L-shaped structure” due to the positioning of the centromere somewhat below the midpoint. This results in one arm being shorter than the other.
Centromere position: Submedial
Arm length ratio: 3.0
Denoted by (sign): smAcrocentric
Acrocentric chromosomes are characterized by one extremely short arm than the other. There are a total of 6 acrocentric chromosomes (chromosome number 13, 14, 15, 21, 22, and Y chromosome) in the human being’s genome. NOTE: An important point to note here is that human males will have a total of 6 acrocentric chromosomes while human females will have only 5 (as females have XX and not XY).
Centromere position: Terminal region
Arm length ratio: 7.0
Denoted by (sign): tTelocentric
Telocentric chromosomes are characterized by the absence of one arm. Since the centromere is present at one end of the chromosome, only one arm is cytologically or microscopic visible. These chromosomes are absent in human beings. Example: house mouse (all chromosomes except Y-chromosome are telocentric)
Centromere position: Terminal sensu stricto
Arm length ratio: ∞
Denoted by (sign): TSubtelocentric
Subtelocentric chromosomes are characterized by two uneven arms since the centromere is placed between the midpoint and the terminal end of the chromosomes. The centromere is closer to the end of the chromosomes.
Centromere position: subterminal
Arm length ratio: 3.1-6.9
Denoted by (sign): st
Figure 2: Different positions of the centromere lead to differently characterized chromosomes. Image Credit: School of Biomedical Sciences WIKI.
Centromere Types
Based on the type of centromere present, there are different terminologies used in current literature. Let’s discuss some of the most common ones.
Acentric
An acentric chromosome lacks a centromere. Since one of the most integral roles played by centromeres is the attachment of spindle fibers during mitosis (mitotic spindle formation) leading to equal distribution of genetic material, acentric chromosomes lead to uneven distribution of genetic material to daughter cells. This can prove lethal for one of the daughter cells.
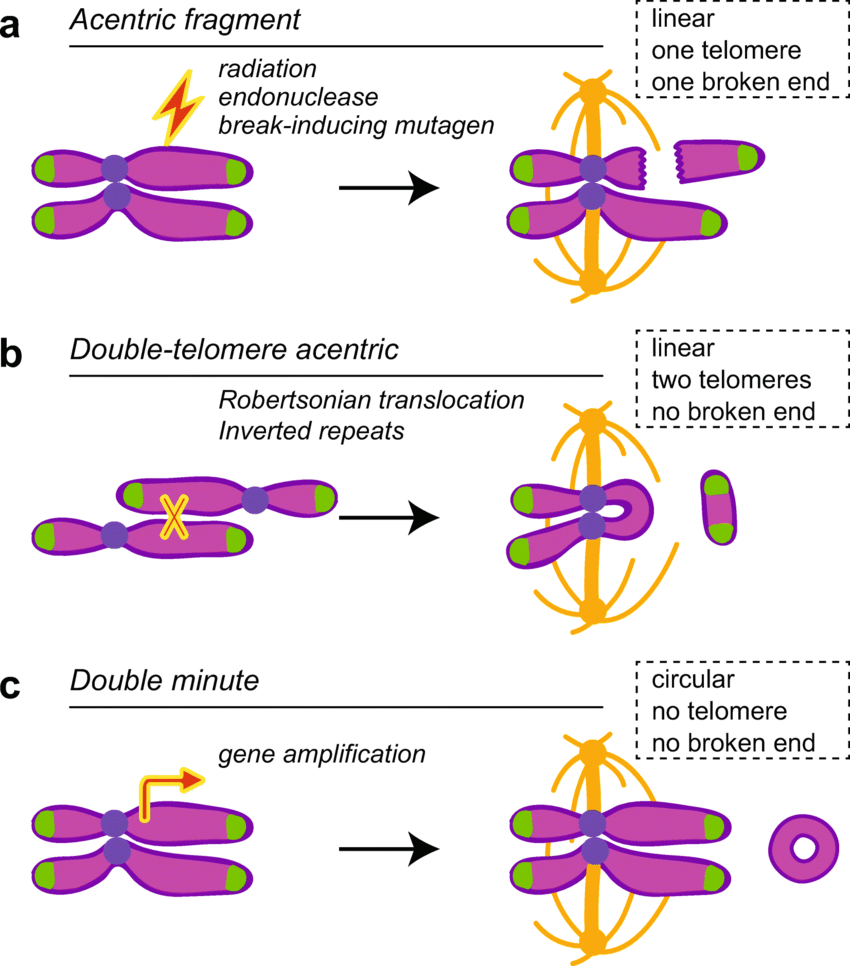
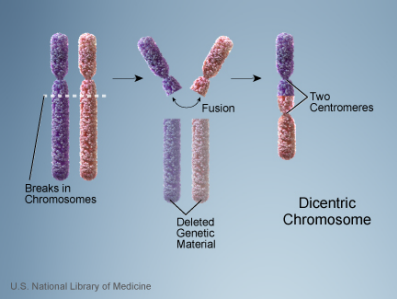
Dicentric
A dicentric chromosome is a chromosome with 2 centromeres. It is abnormal and unstable. Methods of generation of dicentric chromosomes involved one of the following methods:
- Translocation between 2 chromosome segments where each one contains a centromere
- Fusion between 2 chromosome segments where each one contains a centromere
- A Robertsonian translocation
- A paracentric inversion
NOTE: Not all dicentric chromosomes are unstable. Sometimes one of the centromeres gets inactivated (centromere inactivation) leading to only 1 functional centromere. This leads to the formation of a monocentric chromosome that’s capable of normal transmission during cell division.
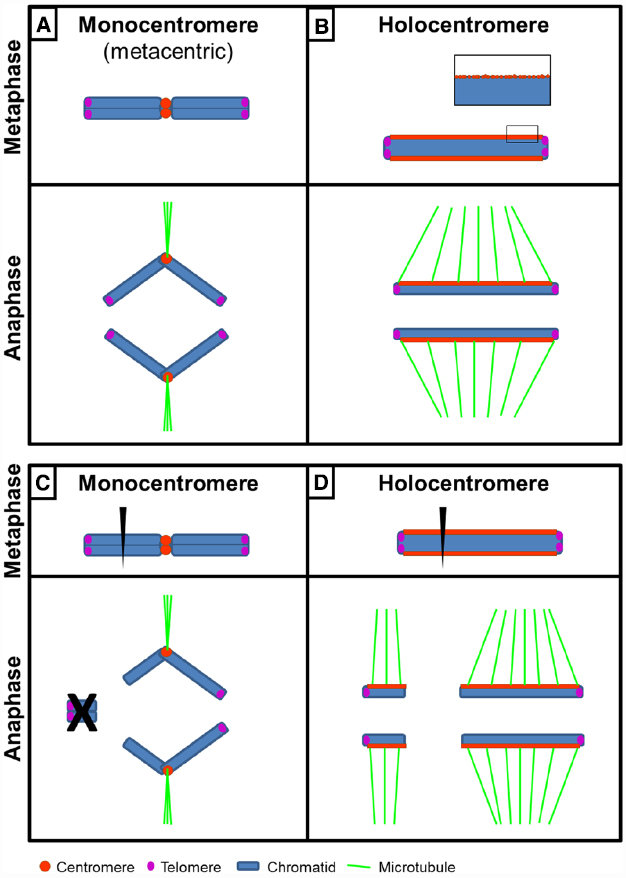
Monocentric
A monocentric chromosome is one with only one centromere in a chromosome. Such a chromosome forms a narrow constriction.
Holocentric
A holocentric chromosome is one with no distinct primary constriction evident at mitosis. This leads to the attachment of spindle fibers along the entire length of the chromosome rather than at a particular centromere point (constriction point). We call the centromeric proteins CENPA and in holocentric chromosomes; these centromere proteins are spread all over the chromosome.
Example: Caenorhabditis elegans (nematode and model organism) is a well-known example of an organism with holocentric chromosomes.
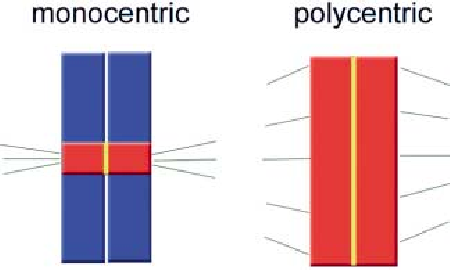
Polycentric
A polycentric chromosome is one with multiple centromeres.
Human chromosomes
Human beings have a pair of 23 eukaryotic chromosomes. The table below describes the type of human chromosomes based on the centromere characteristics:
Table 1: Description of different chromosomes based on centromere type. | |
|---|---|
| Chromosome number | Chromosome type |
| 1 | Metacentric |
| 2 | Submetacentric |
| 3 | Metacentric |
| 4 | Submetacentric |
| 5 | Submetacentric |
| 6 | Submetacentric |
| 7 | Submetacentric |
| 8 | Submetacentric |
| 9 | Submetacentric |
| 10 | Submetacentric |
| 11 | Submetacentric |
| 12 | Submetacentric |
| 13 | Acrocentric |
| 14 | Acrocentric |
| 15 | Acrocentric |
| 16 | Metacentric |
| 17 | Submetacentric |
| 18 | Submetacentric |
| 19 | Metacentric |
| 20 | Metacentric |
| 21 | Acrocentric |
| 22 | Acrocentric |
| 23-X | Submetacentric |
| 23-Y | Acrocentric |
Data Source: Akanksha Saxena of Biology Online
Sequence
The DNA sequence of the centromere and its importance depends on the type of centromere. There are essentially 2 types of centromeres; regional and point centromeres.
- Regional Centromere: The sequence of regional centromeres reveals a large amount of repetitions in the DNA (repetitive DNA/satellite DNA). Alpha satellite DNA, also known as alphoid DNA is the centromeric DNA repeats/units in human centromeres. Although the sequence of regional centromeres contributes to the centromere function, it is not definitive. Most of the DNA in regional centromeres is packaged into heterochromatin form (an inactive form of DNA). The sequence of repetitive DNA in the regional centromeres is similar but not exactly identical. The assembly of this type of centromere is majorly dependent on epigenetic factors and not on the sequence.

Figure 7: Eukaryotic centromeres. Different-sized DNA sequences of centromeric DNA in different eukaryote species. One can notice the “point centromere” of a small size (125bp) in Saccharomyces cerevisiae (budding yeast) and the “regional centromere” of a size as huge as 0.2-5Mb in Homo sapiens (human beings). Image Credit: K.M. Stimpson. - Point Centromere: Centromeric DNA sequence of the point centromere is integral to the centromere identity and function. This type of centromere is smaller in size and more compact.
Inheritance
Centromeric identity is inherited by the means of epigenetics and not DNA sequence (as noted in metazoans).
The assembly of daughter chromosomes’ centromere is determined by the pattern of parent chromosomes’ centromere; thus indicating the key determinant to be “epigenetic control”.
CENP-A is a histone H3 variant. It is proposed to be the epigenetic marker for centromeres.
There is still no clarity or research on the original method by which centromere is initially specified after which it’s subsequently propagated by epigenetic means.

Structure
State of centromere DNA: Heterochromatin
Why is the heterochromatin state most suitable for centromeric DNA? This is so because it aids the recruitment of the cohesin complex. The Cohesin complex is the structural framework that mediates the sister chromatid cohesion after DNA replication. It also coordinates the sister chromatid separation during anaphase.
CENP-A
CENP-A plays a major role in this process as it is the centromere-specific variant of histone H3 protein in humans.
CENP-A ensures that the kinetochore assembly is carried out efficiently on the centromere.
Islands of retroelements are the most integral components of centromeres in Drosophila melanogaster (fruitflies).
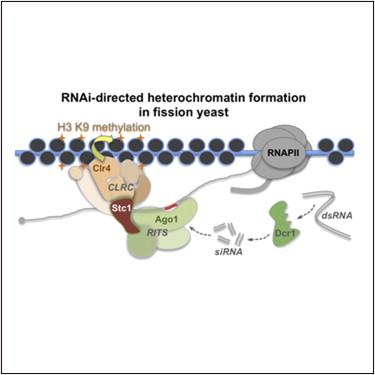
RNA interference
The process of RNAi (RNA interference) is quite important in the formation of centromeres in Schizosaccharomyces pombe (fission yeast).
Centromeric Aberrations
Neocentromeres: Centromeres at novel sites on chromosomes. This results from the repositioning of the centromere. This phenomenon has been reported from 20 different chromosomes to date in human clinical studies.
Spontaneous formation of neocentromeres on fragmented chromosomes.
Dysfunction and Disease
Several disorders and diseases are related to the deregulation of centromere assembly. Studies have shown that anomalies in centromere protein targeting can lead to cancer. A study by Tomonaga, T. et al. reported that overexpression and mistargeting of CENP-A in human beings led to primary colorectal cancer. (Ref-7)
There are several other studies that have provided evidence showing the potential linkage of erroneous centromeric assembly with cancers. (Ref-8,9,10)
Some tumors (tumor cells) have been linked to aberrant DNA methylation in the centromeric region and it is also linked to aneuploidy and genomic instability. (Ref-11)
Repair of centromeric DNA
Homologous recombination repair ensures the repairs of centromeric breaks. This is done throughout the cell cycle. This ensures that no inaccurate mutagenic DNA repair pathways are activated. This also ensures the preservation of centromeric integrity.
The curious case of Acrocentric Chromosomes
The shorter arm (p-arm) of the acrocentric chromosomes is significantly smaller than q-arm, thereby containing relatively very less genetic material. This makes the translocation via a balanced Robertsonian translocation easier and quite spontaneous in the chromosome. The acrocentric chromosomes not only contain protein-coding genes but are also NORs (nucleolus organizer regions).
NORs are important for the transcription of the rRNA (ribosomal RNA). In cases of unbalanced translocations in acrocentric chromosomes, a number of diseases are manifested. Moreover, unbalanced translocations are “more common in acrocentric chromosomes” than non-acrocentric chromosomes.
Examples of some commonly known diseases and disorders linked to Robertsonian translocation in the acrocentric chromosomes are:
- Down’s Syndrome: Involves chromosome number 21
- Patau Syndrome: Involves chromosomes number 13

Take the Centromere – Biology Quiz!
References
- Lejeune J, Levan A, Böök JA, Chu EHY, Ford CE, Fraccaro M, Harnden DG, Hsu TC, Hungerford DA, Jacobs PA, et al. 1960. A proposed standard system of nomenclature of human mitotic chromosomes. The Lancet 275: 1063–1065.doi:10.1016/S0140-6736(60)90948-X
- Antonarakis, S. E. (2022). Short arms of human acrocentric chromosomes and the completion of the human genome sequence. Genome Research, 32(4), 599-607.
O’Connor, C. (2008) Human chromosome translocations and cancer. Nature Education 1(1):56 - Gernand, D., Demidov, D., & Houben, A. (2003). The temporal and spatial pattern of histone H3 phosphorylation at serine 28 and serine 10 is similar in plants but differs between mono-and polycentric chromosomes. Cytogenetic and genome research, 101(2), 172-176.
- Zasadzińska, E., Huang, J., Bailey, A. O., Guo, L. Y., Lee, N. S., Srivastava, S., … & Foltz, D. R. (2018). Inheritance of CENP-A nucleosomes during DNA replication requires HJURP. Developmental cell, 47(3), 348-362.
- Bayne, E. H., White, S. A., Kagansky, A., Bijos, D. A., Sanchez-Pulido, L., Hoe, K. L., … & Allshire, R. C. (2010). Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell, 140(5), 666-677.
- Tomonaga, T., Matsushita, K., Yamaguchi, S., Oohashi, T., Shimada, H., Ochiai, T., … & Nomura, F. (2003). Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer research, 63(13), 3511-3516.
- Sharma, A. B., Dimitrov, S., Hamiche, A., & Van Dyck, E. (2019). Centromeric and ectopic assembly of CENP-A chromatin in health and cancer: old marks and new tracks. Nucleic Acids Research, 47(3), 1051-1069.
- Oka, N., Kasamatsu, A., Endo-Sakamoto, Y., Eizuka, K., Wagai, S., Koide-Ishida, N., … & Uzawa, K. (2019). Centromere protein N participates in cellular proliferation of human oral cancer by cell-cycle enhancement. Journal of Cancer, 10(16), 3728.
- Sun, X., Clermont, P. L., Jiao, W., Helgason, C. D., Gout, P. W., Wang, Y., & Qu, S. (2016). Elevated expression of the centromere protein‐A (CENP‐A)‐encoding gene as a prognostic and predictive biomarker in human cancers. International journal of cancer, 139(4), 899-907.
- Scelfo, A., & Fachinetti, D. (2019). Keeping the Centromere under Control: A Promising Role for DNA Methylation. Cells, 8(8), 912. https://doi.org/10.3390/cells8080912
©BiologyOnline.com. Content provided and moderated by Biology Online Editors.