Epistasis
n., plural: epistases
[ɪˈpɪstəsɪs]
Definition: The modified expression of a gene by an unrelated gene
Table of Contents
Epistasis Definition
What is epistasis in genetics? How does epistasis occur? The epistasis definition, in biology, refers to a situation in which the expression of one or more other genes modifies (e.g., masks, inhibits, or suppresses) the expression of a particular gene ratio. When we define epistasis based on its roots, it’s interesting that the Greek roots of the word “epistasis” indicate “standing upon“.
Epistasis is a phenomenon in genetics whereby the presence or absence of mutations in one or more extra genes, referred to as modifier genes (genetic modifiers), affects the outcome of a gene mutation. In other words, the mutation’s fate is determined by the genetic environment in which it occurs. Hence, the epistatic effect or epistatic mutations act differently on their own than when they combine.
The original definition of the term “epistasis” was the phenomenon in which the influence of a gene variant (genetic variants) is overridden by that of another gene.
The concept of epistasis was first presented in the study of genetics in 1907 and is currently used in biochemistry, computational biology, and evolutionary biology. The phenomenon results from interactions between or within genes, which produce non-linear effects. For example, mutations in regulators of gene expression are required, as are many mutations before a gene loses its function. This can put the organism at complex disease and general disease risk.
Epistasis has a significant impact on how evolutionary landscapes are shaped, which has significant ramifications for both evolution and the evolvability of phenotypic features.
The term “epistasis” in genetics refers to the interaction of genes at two or more loci, and as a result, the presence of one or more modifier genes affects the function of the gene. A gene or allele in the interaction may hide the phenotypic expression of the other genes or alleles. Such a gene or allele masking the effect is referred to as epistatic. In contrast, the other gene(s) or allele(s) being masked is/are referred to as hypostatic. An example of epistasis is the fur color of Labrador retrievers, which is a polygenic trait. Two genes are interacting to determine its fur color. One gene (represented by B) determines the fur color whereas the other gene (represented by E) controls the expression of the gene B. The presence of homozygous recessive alleles ee would mask gene B (regardless of the dominant B alleles). This is an example of epistasis, particularly the recessive type.Etymology: Greek epístasis (stopping, stoppage)
History
The definition and usage of the term “epistasis” have evolved significantly during the course of genetics history. The phrase was first coined by William Bateson along with his friends Florence Durham and Muriel Wheldale Onslow. Early natural selection models developed in the early 20th century assumed that each or one gene uniquely contributed to fitness against a background of average other genes.
Population genetics is still taught in some introductory courses in this manner. Evolutionary geneticists tend to view epistasis as the exception due to the way population genetics was created. Yet, the expression of any one allele typically involves complicated interactions with a number of other alleles.
Gene A is epistatic and gene B is hypostatic in classical genetics if genes A and B (two genes – two separate genes) are mutated and each mutation alone results in a distinct phenotype but the two mutations combined create the same phenotype as the gene A mutation. For instance, the gene for brown hair is epistatic to the gene for total baldness.
In this sense, genetic dominance, the interaction of alleles at the same gene locus, can be compared with epistasis. Epistasis began to be researched in relation to polygenic inheritance and epistasis as well as quantitative trait loci (QTL) as the field of human molecular genetics, quantitative genetics and molecular biology advanced.
Nowadays, it is normal practice to measure the amplitude of a phenotypic ratio (such as height, color, or growth rate) or to measure the activity of proteins biochemically in order to quantify the impact of genes (e.g. binding or catalysis).
The goal of increasingly complex computational and evolutionary biology models is to explain how epistasis affects evolution on a genome-wide scale. Some studies attempt to prioritize epistatic couples since it is difficult to identify epistatic gene pairs both computationally and statistically.
Types of Epistasis
The six types of epistasis gene interactions include dominant, dominant inhibitory, duplicate dominant epistasis, duplicate recessive, polymeric gene interaction, and recessive.
Dominant epistasis ratio, also known as simple epistasis, occurs when a dominant epistatic allele at one location conceals the expression of both dominant and recessive alleles at another, one locus.
Recessive epistasis refers to the masking of an expression by a recessive allele. By suppression, some genes can also cover up other genes or a version of a gene. Since the gene is acting as a suppressor—a factor that prevents the production of another allele—this is known as dominant inhibitory or suppression epistasis.
Epistasis with duplicate types is dependent on two genetic loci. The loci definition in biology is “the actual location of a particular gene on a chromosome”.
Double dominant epistasis pathway, also referred to as duplicate gene action, occurs when a dominant allele conceals the expression of recessive alleles at two loci.
Duplicate recessive epistasis occurs when a recessive allele conceals the expression of dominant alleles at two loci. Because both genes are necessary for the right phenotype to exist, it is often referred to as complementary gene action.
Polymeric gene interaction is the result of two dominant alleles combining to increase the phenotype or generate a median variance. Each dominant allele alone generates physical epistatic traits distinct from the sum of the dominant alleles. With only two dominant alleles, this results in three phenotypes. No dominant allele is surpassing the other dominant allele as a result.
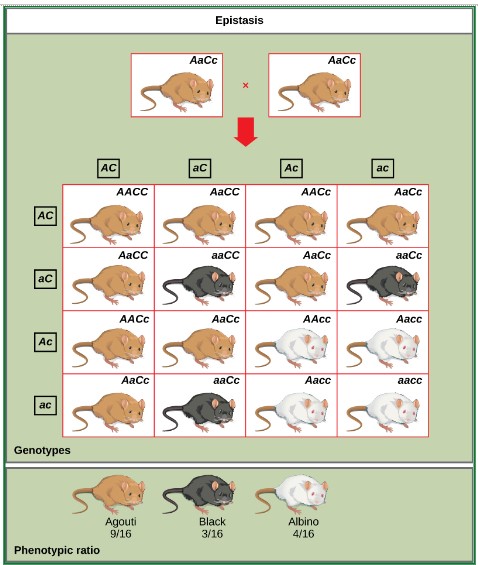
Classification
Epistasis terminology varies throughout scientific disciplines. When a mutation is implicitly harmful, geneticists frequently discuss the wild-type and mutant alleles as well as genetic enhancement, synthetic lethality, genetic background, and genetic suppressors.
A biochemist, on the other hand, might prefer to concentrate more on advantageous mutations and, as a result, employ phrases like reciprocal sign epistasis and compensatory mutation to explicitly describe the consequence of a mutation.
Moreover, there are variations between epistasis within a haploid or diploid genome and epistasis inside a single gene (biochemistry) (genetics). Epistasis is a term used to describe when the effects of various genetic loci are no longer considered to be “independent”.
Because different fields of biology understand “independence” in different ways, confusion frequently results. The descriptions that follow make an effort to cover the various terminology and their relationships.
Additivity
If the combined effects of the two mutations equal the effects of the single mutations, then two mutations are regarded as being entirely additive genetic variance. This happens when genes do not communicate with one another, for instance, when they act through various metabolic pathways.
Simply said, additive features have been researched throughout the history of genetics, but they are very uncommon now because most genes show at least some degree of epistatic interaction.
Magnitude epistasis
Positive epistasis occurs when the phenotype of the double mutation is fitter than would be predicted from the effects of the two single mutations. Positive epistasis between advantageous mutations leads to higher functional benefits than anticipated. Positive epistasis between harmful mutations guards against the adverse consequences to result in a less drastic decline in fitness.
Negative epistasis, on the other hand, occurs when two mutations work together to produce a phenotype that is less fit than would be predicted from their individual impacts. Negative epistasis between advantageous mutations results in fitness gains that are less pronounced than anticipated, and negative epistasis between harmful mutations results in fitness losses that are more pronounced than intended.
Sign epistasis
Synergistic epistasis is the term used when the combined effects of two mutations on fitness are more extreme than would be predicted from their individual effects. It is known as antagonistic epistasis when the fitness difference between the double mutant and the wild type is less than would be predicted from the impact of the two single mutations.
Negative epistasis is therefore also synergistic for harmful mutations, whilst positive epistasis is antagonistic; on the other hand, positive epistasis is synergistic for individual beneficial mutations, but negative epistasis is antagonistic.
When a double (deleterious) mutant manifests a more severe phenotype than the combined effects of the single mutants, the phrase genetic enhancement is occasionally employed. Some creationists refer to strong positive epistasis as irreducible complexity (although most examples are misidentified) and signs of strong genes.
Haploid organisms
We might think of several types of epistasis as changing the degree of a phenotype following mutation independently (Ab and aB) or in combination (AB) in a haploid organism having genotypes (at two loci) ab suppressor, Ab, aB, or AB (AB).

Diploid organisms
Epistasis is made more difficult in diploid organisms since each gene is present in two copies. Among heterozygotes, interactions between each locus’s two copies are also possible in addition to epistasis. There are eight distinct types of gene interaction for a two locus, two allele system.
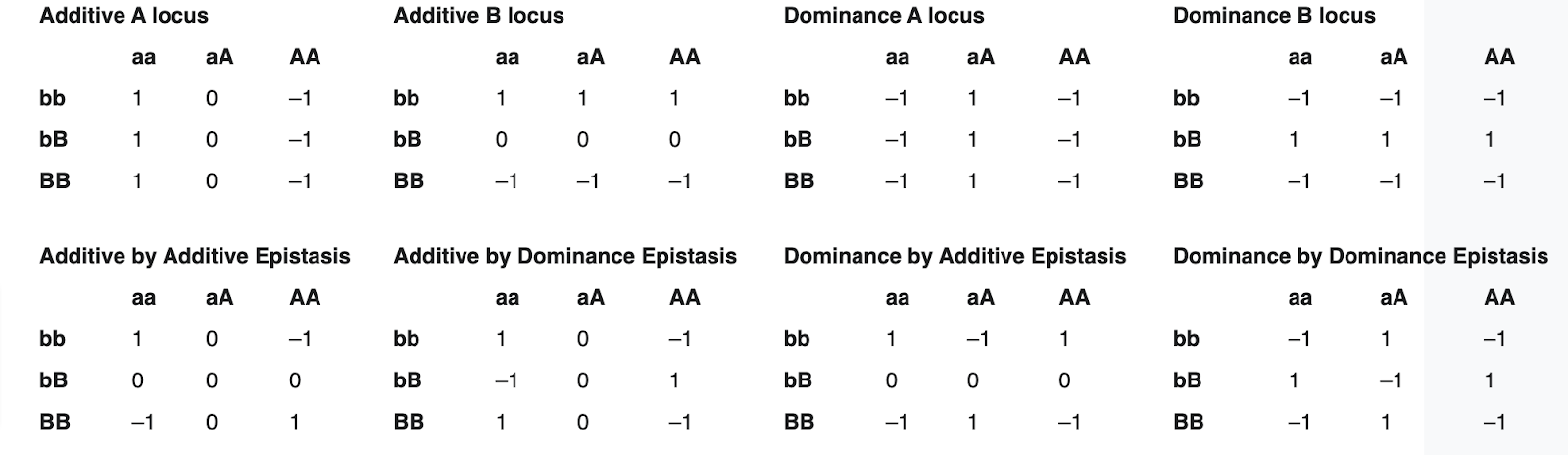
Genetic and Molecular Causes
Let’s next learn about the genetic and molecular causes of epistasis.
Additivity
When several genes work simultaneously to produce the same result, this may be the case. For instance, several enzymes that break down various phosphorylated elements from the environment may function additively when an organism needs phosphorus to increase the quantity of phosphorus available to the organism.
But, phosphorus will eventually stop being the limiting factor for growth and reproduction, and as a result, additional increases in phosphorus metabolism will have less or no effect (negative epistasis).
Also, it has been particularly discovered that some combinations of gene mutations are additive. In light of the fact that most genes interact with hundreds or thousands of other genes, it is now believed that strict additivity is the exception rather than the rule.
Epistasis between genes
The interactions between the genes inside the genomes of organisms lead to epistasis. If the genes produce proteins that, for example, are independent parts of a multi-component protein (like the ribosome), inhibit each other’s activity, or alter the other gene’s protein, then this contact may be direct (such as by phosphorylation).
If the genes encode parts of a metabolic process or network, developmental pathway, signaling pathway, or transcription factor network, the interaction may also be indirect. Without the enzymes that produce the essential precursors in the metabolic pathway, the gene encoding the enzyme that synthesizes penicillin, for instance, is useless to a fungus.
Epistasis within genes
Two codons inside a gene may also be non-additive, just as two different genes might be if they interact. When a harmful mutation can be offset by a different mutation occurring inside the same gene, this phenomenon is commonly referred to as intragenic suppression in epistasis genetics.
The rIIB cistron (gene) mutations in bacteriophage T4 mutants were examined, and it was found that some paired combinations of mutations could suppress one another, giving rise to more closely resembling wild-type phenotypes in the double mutants than in either mutant alone.
Genetic recombination data was used to determine the linear map order of the mutants. From these data, the triplet nature of the genetic code was logically determined for the first time in 1961, as well as other important characteristics of the code.
Furthermore, intragenic suppression can happen when a protein’s amino acids interact. Additive mutations are uncommon because of the complexity of protein functions like folding and action.
A dispersed internal network of cooperative connections maintains the tertiary structure of proteins (hydrophobic, polar, and covalent). When one mutation modifies the immediate environment of another residue, epistatic interactions happen (either by directly contacting it or by inducing changes in the protein structure).
For instance, in a disulfide bridge, the presence of a second cysteine at the proper place causes the two cysteines to create a chemical connection that increases the protein’s stability. Until then, a single cysteine does not influence the stability of the protein.
The double-cysteine variation had a significantly higher stability than any of the single-cysteine versions, and this was seen as positive epistasis. A few essential amino acids are positioned into exact geometries by the protein structure of enzymes to create an active site that can carry out chemical reactions.
Since these active site networks usually require the collaboration of numerous components, changing any one of them severely impairs activity; nevertheless, changing a second component only little affects the enzyme’s already-inactivated state.
For instance, eliminating any one of the many enzymes that make up the catalytic triad will cause activity to drop to levels so low that the organism is no longer viable.
Heterozygotic epistasis
Each gene is present in two copies in diploid organisms. The two separate copies of the allele may interact with one another to generate epistasis if these are different (heterozygous/heteroallelic). Sometimes, this is referred to as interallelic complementation or allelic complementation.
Transvection, when an enhancer from one allele acts in trans to trigger transcription from the promoter of the second allele, is one mechanism that could be responsible. Conversely, two non-functional RNA molecules may be trans-spliced to create a single, functional RNA.
Similar to this, proteins that operate as dimers may produce a heterodimer made up of one protein from each alternative gene, and this heterodimer may exhibit distinct properties from the homodimer of one or both variations.
During a mixed infection, two bacteriophages T4 mutants that are deficient at various points in the same gene may experience allelic complementation. In other words, each mutant cannot create viable offspring when it is infected alone; nevertheless, when two complementing mutants are infected together, viable phages are produced.
For several genes that encode bacteriophage structural proteins, intragenic complementation has been shown, demonstrating that these proteins operate as dimers or even higher-order multimers.
Watch this vid about epistasis and other forms of Non-Mendelian Inheritance:
Note it!
One gene frequently masks or interferes with the expression of another gene in epistasis (antagonistic epistasis). When distinct genes are a part of the same metabolic process, epistasis frequently happens. A gene product in the same metabolic pathway may be necessary for the expression of the gene.
Examples of Epistasis
Here are some examples of epistasis:
Labrador Retriever
The coat color of the well-known Labrador retriever dog breed is a good illustration of epistasis. Only the black and chocolate coat color genes are found in labrador retrievers. Yet in the dog park, you see yellow Labrador retrievers running around. This happens because so-called “extension genes” , which are recessive epistatic genes, prevent color pigment from actually getting to the fur.

Epistasis examples in humans
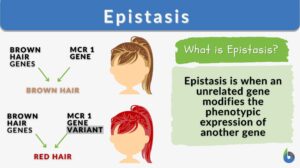
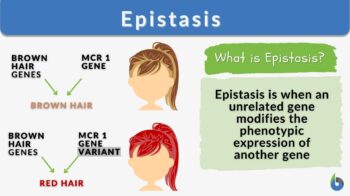
Certain polygenic characteristics, like skin, eye, and hair color, necessitate the existence of an epistatic gene. However, in human populations, not all polygenic traits are the outcome of epistasis. For instance, the existence of an epistatic gene is not necessary for the development of blonde, light-brown, or dark-brown hair. Yet, red hair cannot be expressed in humans without an epistasis gene.
Yet, not all epistasis cases are as benign as red hair. Some epistasis examples, such as albinism, have more significant phenotypic outcomes. In either case, a deeper understanding of the genetic interactions that cause physical features such as albinism or red hair would aid in comprehending the phenomenon of epistasis.
Several genes work together to determine the polygenic trait of hair color. For instance, a lot of a pigment called eumelanin would have to be present for a human to have dark-brown hair or hair that appears black as a physical feature. However, the generation and distribution of eumelanin along a hair strand are regulated by a number of genes.
Tyrosine, an amino acid, undergoes numerous chemical interactions or transformations to produce eumelanin. One of these modifications is brought on by the melanocortin 1 receptor (MCR1) gene. The MCR1 gene produces the MCR1 protein, which combines with tyrosine to produce eumelanin, which is then synthesized.
There are at least 10 more genes that affect hair color. OCA2, which contributes to the synthesis of the P protein necessary for the creation of melanin, and HERC2, a protein-coding gene involved in the binding of proteins that results in pigments, are two other instances.
Unfortunately, not all hair color genes have been mapped to the point that researchers are certain of the precise functions of each gene. The correlation between genes and physical features provides a broad understanding of what genes are responsible for.
Nonetheless, the epistatic gene responsible for the physical attribute of red hair color in humans has been identified. An accumulation of a pigment called pheomelanin is required for someone to get red hair. This pigment is involved in tyrosine’s transformation. Pheomelanin is consumed in a chemical reaction that yields eumelanin if the MCR1 protein is added. However, a variant of MCR1 gene was found to prevent the conversion of pheomelanin into eumelanin, resulting in the buildup of pheomelanin, which is observed in red hair phenotype.
Combs in Chickens
Miko chicken research brought about further depth into British geneticists William Bateson and R. C. Punnett‘s studies in the first decade of the 20th century. It demonstrated that the interplay of two distinct genes in chickens was responsible for the shape of their combs. The characteristic combs that each variety of chicken has were known to Bateson and Punnett. For example, Leghorns have a “single” comb, Brahmans have a “pea” comb, and Wyandottes have a “rose” comb. These can only result if true breeding occurs.
The Wyandotte chicken also comes in multiple colours as a result of genetic epistasis. The colours range from regular whites, browns and blacks but the purple Wyandotte chicken colors are a bit rare and truly a sight to behold.

Take the Epistasis – Biology Quiz!
References
- Azevedo, R. B. R., Lohaus, R., Srinivasan, S., Dang, K. K., & Burch, C. L. (2006). Sexual reproduction selects for robustness and negative epistasis in artificial gene networks. Nature, 440(7080), 87–90. https://doi.org/10.1038/nature04488
- Chattopadhyay, S., Thomsen, H., Yadav, P., da Silva Filho, M. I., Weinhold, N., Nöthen, M. M., Hoffman, P., Bertsch, U., Huhn, S., Morgan, G. J., Goldschmidt, H., Houlston, R., Hemminki, K., & Försti, A. (2019). Genome-wide interaction and pathway-based identification of key regulators in multiple myeloma. Communications Biology, 2, 89. https://doi.org/10.1038/s42003-019-0329-2
- Definition of locus—NCI Dictionary of Genetics Terms—NCI (nciglobal,ncienterprise). (2012, July 20). [NciAppModulePage]. https://www.cancer.gov/publications/dictionaries/genetics-dictionary/def/locus
- Domingo, E., Sheldon, J., & Perales, C. (2012). Viral quasispecies evolution. Microbiology and Molecular Biology Reviews: MMBR, 76(2), 159–216. https://doi.org/10.1128/MMBR.05023-11
- Epistasis | Biology for Majors I. (n.d.). Retrieved April 8, 2023, from https://courses.lumenlearning.com/wm-biology1/chapter/reading-epistasis-2/
- Epistasis and Its Effects on Phenotype | Learn Science at Scitable. (n.d.). Retrieved April 8, 2023, from http://www.nature.com/scitable/topicpage/epistasis-gene-interaction-and-phenotype-effects-460
- Epistasis—Alchetron, The Free Social Encyclopedia. (2017, August 18). Alchetron.Com. https://alchetron.com/Epistasis
- Gene Interactions and Epistasis | Biology | JoVE. (n.d.). Retrieved April 8, 2023, from https://www.jove.com/science-education/10779/epistasis
- Greenfire Farms—Lavender Wyandotte. (n.d.). Retrieved April 8, 2023, from https://greenfirefarms.com/lavender_wyandotte.html
- Gros, P.-A., Le Nagard, H., & Tenaillon, O. (2009). The Evolution of Epistasis and Its Links With Genetic Robustness, Complexity and Drift in a Phenotypic Model of Adaptation. Genetics, 182(1), 277–293. https://doi.org/10.1534/genetics.108.099127
- He, X., Qian, W., Wang, Z., Li, Y., & Zhang, J. (2010). Prevalent positive epistasis in Escherichia coli and Saccharomyces cerevisiae metabolic networks. Nature Genetics, 42(3), 272–276. https://doi.org/10.1038/ng.524
- Karki, P. (2022, September 6). Epistasis- Definition, Classes, Types, Examples, Significances. https://thebiologynotes.com/epistasis-classes-types-examples-significances/
- Kaznatcheev, A. (2019). Computational Complexity as an Ultimate Constraint on Evolution. Genetics, 212(1), 245–265. https://doi.org/10.1534/genetics.119.302000
- Piriyapongsa, J., Ngamphiw, C., Intarapanich, A., Kulawonganunchai, S., Assawamakin, A., Bootchai, C., Shaw, P. J., & Tongsima, S. (2012). iLOCi: A SNP interaction prioritization technique for detecting epistasis in genome-wide association studies. BMC Genomics, 13 Suppl 7(Suppl 7), S2. https://doi.org/10.1186/1471-2164-13-S7-S2
- Szendro, I. G., Schenk, M. F., Franke, J., Krug, J., & de Visser, J. A. G. M. (2013). Quantitative analyses of empirical fitness landscapes. Journal of Statistical Mechanics: Theory and Experiment, 2013(01), P01005. https://doi.org/10.1088/1742-5468/2013/01/P01005
- Weinreich, D. M., Watson, R. A., & Chao, L. (2005). Perspective: Sign epistasis and genetic constraint on evolutionary trajectories. Evolution; International Journal of Organic Evolution, 59(6), 1165–1174.
©BiologyOnline.com. Content provided and moderated by Biology Online Editors.