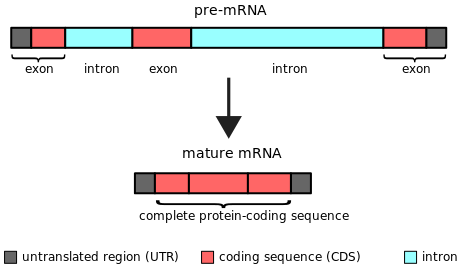
Exon
n., plural: exons
[ˈɛksɑn]
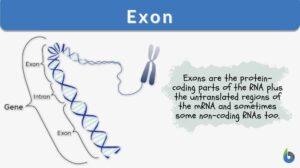
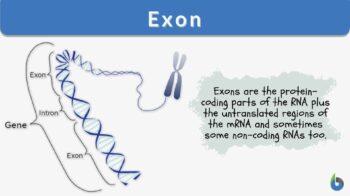
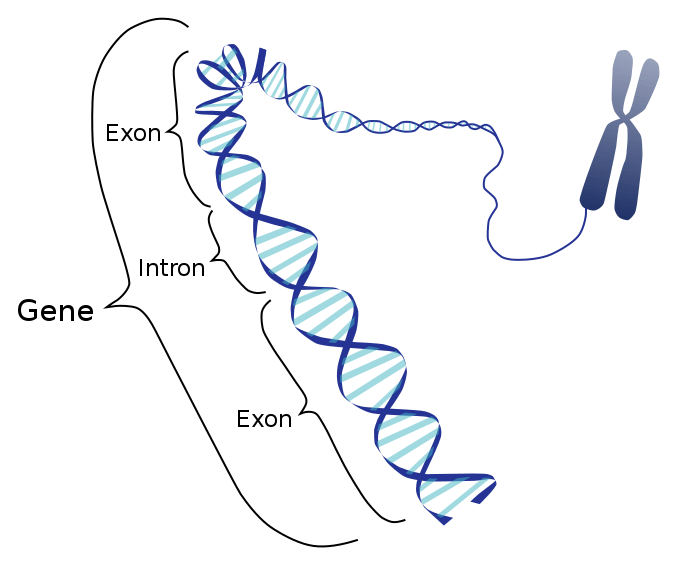
Definition: the protein-coding parts of the RNA plus the untranslated regions of the mRNA
Table of Contents
When we look at the genome sizes and the organism complexity, it becomes quite confusing at times. This is because we encounter some organisms with very small genome sizes like Mycoplasma genitalium (the smallest known genome size in the biological world). Nevertheless, these organisms possess a very complex life cycle and biological interactions. There have been reports of this bacterium becoming resistant to so many antibiotics in 2021. Since this is a disease-causing bacterium (causes STDs), it calls for newer diagnostic options and treatment methods. Studying the genome becomes much more important here on.
So, when we look at the genomes of different organisms, we find them different at many levels. But the basic structure remains the same. “Genome” is the sum total of all the genetic material that an organism has. Within the genome there are 3 components:
- Coding DNA
- Non-coding DNA
- Untranslated regions (UTRs)
When questioned “what is an exon”, we can explain that the ‘coding DNA plus the untranslated regions’, collectively form the “exon” of the genome. On the other hand, ‘non-coding regions of DNA’ are called “DNA introns” of the genome.
Let’s move forward and learn more about exons in genes under the specific headings.
Exon Definition
We can describe the concept of exons from different perspectives.
Biology definition:
- When describing from the ‘DNA point of view’, exons are those parts of a gene that eventually get incorporated in the final structure of a mature RNA post-splicing.
- When describing from the ‘RNA point of view’, exons are protein-coding parts of the RNA plus the untranslated regions of the mRNA (UTR exon mRNA) plus some part that’s actually the non-coding RNA.
Compare: intron
NOTE:
When we are asked to define exon, we tend to usually restrict ourselves to the coding part of the gene. (Common perception: the coding regions of DNA are called exons). We need to note that exons can also be the untranslated regions (UTR- both 5’ UTR and 3’ UTR) and non-coding RNAs (ncRNA; parts of genes that are transcribed from DNA to RNA but aren’t translated ahead from RNA to proteins). So, ncRNA is that part of the exon that undergoes exon transcription, but not translation).
Watch this vid to know what introns and exons are.
History
Exon term, in general, has been used for the ‘expressed region” while intron term is used for intragenic region. It was Walter Gilbert, a well-known biochemist who coined the term ‘exon’. He explained how the introns interrupt and alternate between the consecutive exons. He emphasized how introns are carried along from DNA to pre-mRNA, but then excised off via the “splicing process”, leaving the mature mRNA with only exons to be translated.
Exon Theory of Genes
Walter Gilbert was the one who proposed the “exon theory of genes”. In this, he explained the less transitioning and more permanence in the nature of exons in the gene structure. He explained how in comparison of one gene from different species that are long and enough separated by a sufficient evolutionary distance, one can notice that exons of homologous genes transition very slowly. On the other hand, introns drift quickly and are less conserved since they don’t code for amino acids.

Contribution to Genomes and Size Distribution
Exons make a negligible contribution to the genome if compared to the introns. Although both prokaryotes and eukaryotes possess introns, their presence is more prevalent in the eukaryotes. The records of identification of introns were so few in the prokaryotic genome in the early years of genomics that the idea of “prokaryotes lack introns and have only exons” was widely popularized.
If you notice the genomes of eukaryotes like humans, exons form the minimal part of the whole genome (merely 1.1%), the rest is introns (nearly 24%) or intergenic DNA (nearly 75%) Venter J.C.; et al. (2000). According to an approximation, human genes have 8.8 exons per gene. (Sakharkar M. K., 2004)
Structure and function
Exons comprise both the protein-coding sequence and the untranslated regions (5′ and 3′ UTRs). As explained earlier, not all exons have the UTR regions. Only the 1st exon includes the 5′-UTR and partial coding sequence. Chances of one exon having both 5′-UTR and 3′-UTR are rare and this occurs only in some genes. As discovered over time, some non-coding RNAs also form a part of exons. So, the non-coding RNA transcripts also possess some exons and introns.
Sometimes, one might be perplexed on what type of RNA contains exons, so now we have clarity about it. Both types of RNA, whether coding or non-coding, have exons.
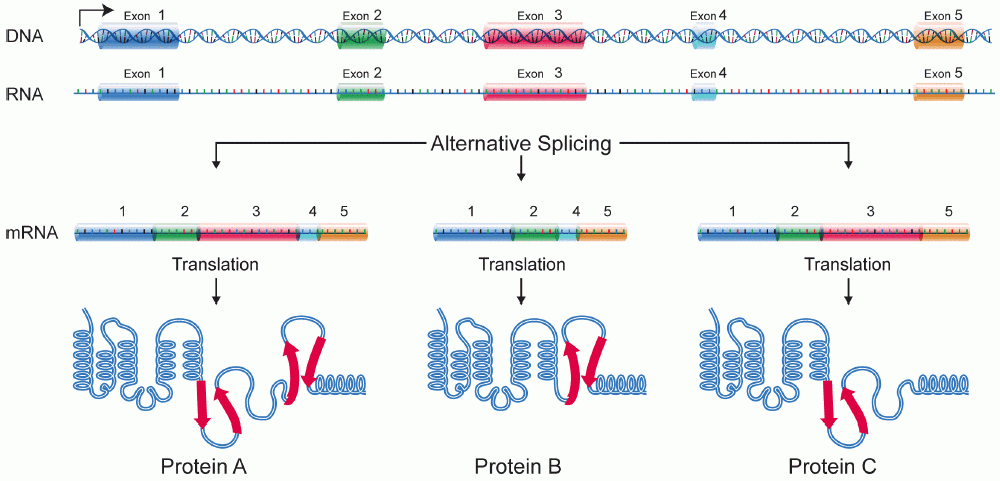
Alternative splicing
The same gene can give rise to different mature mRNAs. This happens by the process of “alternative splicing”. Hence, the mature mRNAs will differ in their exon constitution. These are called alternative exons, which are formed by coming together of different introns.
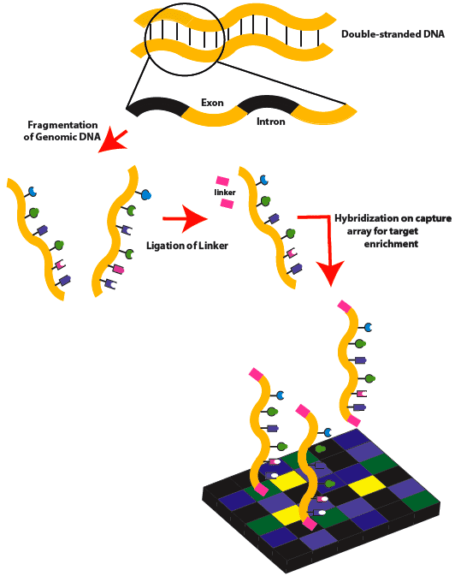
Experimental Approaches Using Exons
Exome sequencing is popularizing as a technique right now. Whole-genome sequencing aids in sequencing the full genome of organisms. But when you require to focus only on the coding part of the genome (or the part that gives rise to functional amino acids and proteins), then ONLY exons need to be sequenced. All the exons together are called “exome”. And sequencing of all the coding regions in the genome is thus called “exome sequencing”.
Interesting Facts about “Exons”
HUMAN GENOME
- Total exons in human genome= 233,785
- Longest exon= 11555 base pair long
- Smallest exon= 2 base pair long
- Largest known gene in human beings= Dystrophin gene (consists of 79 Dystrophin exons)
SMALLEST EXON IN BIOLOGICAL WORLD
The smallest exon reported to date has been recorded to be only 1 nucleotide long. It has been recorded in Arabidopsis genome by Guo Lei and Liu Chun-Ming in 2015. Anaphase Promoting Complex subunit 11 (APC11) gene in Arabidopsis model plant was reported to be carrying a “single-nucleotide exon”.

Try to answer the quiz below to check what you have learned so far about exons.
References
- Glass, J. I., Assad-Garcia, N., Alperovich, N., Yooseph, S., Lewis, M. R., Maruf, M., Hutchison, C. A., 3rd, Smith, H. O., & Venter, J. C. (2006). Essential genes of a minimal bacterium. Proceedings of the National Academy of Sciences of the United States of America, 103(2), 425–430. https://doi.org/10.1073/pnas.0510013103
- Brock J. (2021) Mycoplasma genitalium: A formidable foe in need of new treatment, diagnostic options. Infectious disease news. https://www.healio.com/news/infectious-disease/20211116/mycoplasma-genitalium-a-formidable-foe-in-need-of-new-treatment-diagnostic-options
- Gilbert, W. Why genes in pieces?. Nature 271, 501 (1978). https://doi.org/10.1038/271501a0
- Gilbert W. (1980) DNA SEQUENCING AND GENE STRUCTURE. Nobel lecture, Harvard University, The Biological Laboratories, Cambridge, Massachusetts 02138, USA. https://www.nobelprize.org/uploads/2018/06/gilbert-lecture.pdf
- Venter J.C.; et al. (2000). “The Sequence of the Human Genome”. Science. 291 (5507): 1304–51. doi:10.1126/science.1058040
- Sakharkar, M. K., Chow, V. T., & Kangueane, P. (2004). Distributions of exons and introns in the human genome. In silico biology, 4(4), 387–393.
- Guo Lei, Liu Chun-Ming (2015). “A single-nucleotide exon found in Arabidopsis”. Scientific Reports. 5: 18087. doi:10.1038/srep18087
©BiologyOnline.com. Content provided and moderated by Biology Online Editors.