Induced fit model
n., [ɪnˈduːst fɪt ˈmɑdl̩]
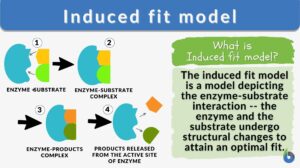
Definition: an enzyme-substrate interaction depicting that both the enzyme and substrate undergo changes to attain an optimal fit
Table of Contents
Induced-Fit Model Definition
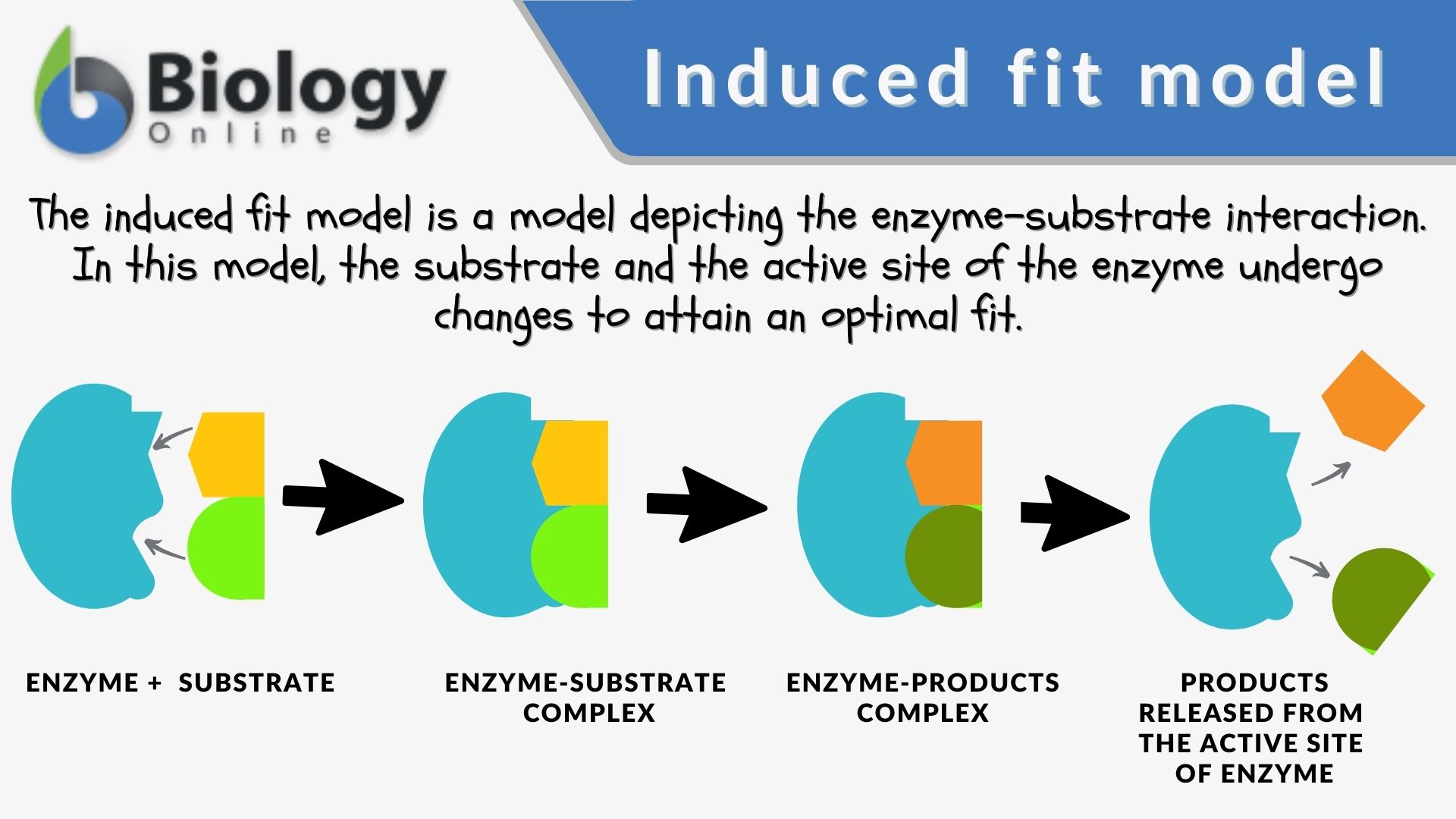
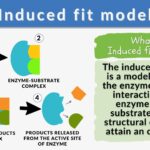
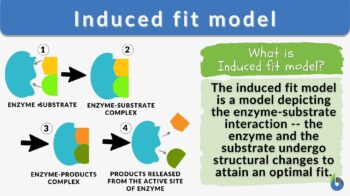
The induced-fit model is a model for enzyme–substrate interaction to depict the dynamic interaction between an enzyme and its substrate. Both the enzyme and the substrate molecules undergo conformational changes to achieve optimal binding and catalytic efficiency. The substrate is capable of inducing the proper alignment of the active site of the enzyme, causing the latter to subsequently perform its catalytic function. It is opposed to the lock-and-key model that is also used to describe the enzyme-substrate interaction. In the induced fit framework, both the substrate and the enzyme change in conformation until the substrate is completely bound to the enzyme, at which point the final shape and charge are determined. The binding induces the enzyme to perform its catalytic function.
Compare: Lock-and-key model See also: enzyme, active site, substrate
Lock-and-key vs. Induced Fit Model
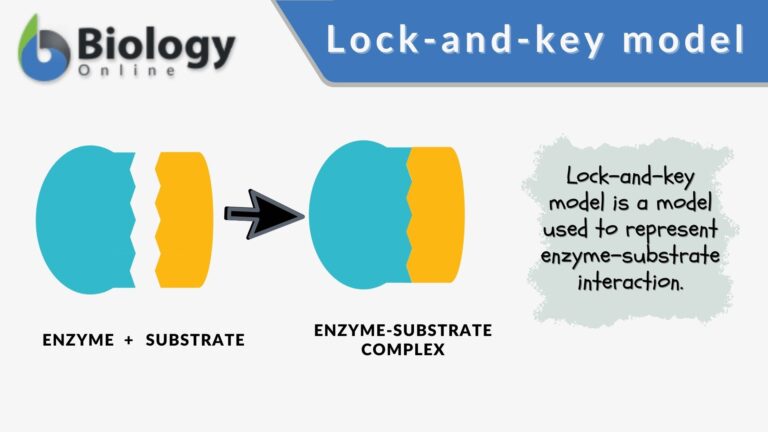
The lock-and-key model is the first to be proposed as it was first postulated by Emil Fischer in 1894 whereas the induced fit model was conceived by Daniel Koshland in 1958. In the lock-and-key model, the interaction of the substrate and the enzyme is likened to a key (the substrate) that is highly specific to the lock (the active site of the enzyme). It depicts a rather static and rigid form of interaction. Unlike the lock-and-key model, the induced fit model proposes that enzymes are rather flexible structures in which the active site continually reshapes by its interactions with the particular substrate until the time the substrate is optimally bound to it. The conformational selection model is the more accepted model for enzyme-substrate complex although the lock-and-key model is vital in laying the groundwork for the understanding of the complex process of enzyme and substrate interactions, including the conceptualization of the induced fit theory. Furthermore, the line that splits the frameworks of the lock and key and the induced fit models is ambiguous and not absolute.
Table 1: Lock-and-key Model vs. Induced Fit Model | ||
|---|---|---|
Lock-and-key Model | Induced Fit Model | |
| Definition | A conceptual framework of the enzyme-substrate interaction where the enzyme and the substrate possess specific complementary geometric shapes that fit exactly into one another | A conceptual framework of the enzyme-substrate interaction in which the enzyme and the substrate undergo conformational changes upon interaction for a more complementary fit |
| Molecular Interaction | Enzyme and substrate fit together like a lock and key, with no significant conformational changes in either molecule | Dynamic interaction where both enzyme and substrate adjust their shapes to achieve optimal binding |
| Specificity | Highly specific, and only substrates with a matching shape can bind to the active site | Specific, but the enzyme and substrate can induce changes in each other’s structures for better compatibility |
| Flexibility | The enzyme’s active site does not undergo significant changes upon substrate binding. | Enzyme’s active site and substrate change to achieve a more optimal fit |
| Analogy | Analogous to a lock and key where the key (substrate) fits precisely into the lock (enzyme). | Analogous to a handshake where both hands adjust to fit together during the interaction |
| Example | One typical example is the interaction of the enzyme chymotrypsin and its substrate peptide that fits precisely into the rigid active site of chymotrypsin (Note: not always absolute as chymotrypsin can also undergo conformational changes when binding with certain peptide substrates) | DNA polymerase enzyme interacting with the DNA nucleotide as its substrate by undergoing conformational changes to accommodate the incoming nucleotide, which may also alter its structure for a better fit |
| Illustration |
|
|
Watch the vid below for the differences between the two models.
Key Components
Components of the Induced Fit Model:
- Enzyme: the enzyme structure is a three-dimensional protein configuration. It has an active site where the substrate can bind or interact and where the catalytic reaction takes place.
- Substrate: a biological molecule with structural features that complement the binding site or the active site of an enzyme during the enzyme-catalyzed reactions.
What the Induce Fit model mean in biological processes and chemical reactions
In the induced fit hypothesis, the enzyme is depicted as a fluid structure as it is not rigid but has a rather flexible nature owing to the presence of the side chains of the amino acids in the active site that often act as catalytic groups. These residues interact with the functional groups on the substrate, helping to stabilize the molecules by orienting them into a specific configuration. The enzyme surface away from the active site also contributes to the enzyme’s catalytic function. During the event of the enzyme-substrate interaction, the conformational change can propagate to the enzyme surface, which then further promotes the overall stability of the enzyme’s structure and function. Furthermore, the binding energy used during the interaction is vital to the overall efficiency of the catalytic process. The structural changes that lead to the optimal binding of the enzyme and the substrate result in a more stable complex. The interaction of the enzyme and the substrate is essential in expediting a chemical reaction, and thus, a biological process could also proceed to completion at a remarkably faster pace.
Examples
Here are examples of enzyme-substrate complexes depicting the induced fit model:
- DNA Polymerase (enzyme) and DNA nucleotides (substrate)
- Hexokinase (enzyme) and glucose (substrate)
- Ribonuclease A (enzyme) and RNA (substrate)
- Acetylcholinesterase (enzyme) and acetylcholine (substrate)
Answer the quiz below to check what you have learned so far about the Induced fit Model.
References
- Seema Anil Belorkar, & Sudisha Jogaiah. (2022). Enzymes—past, present, and future. Elsevier EBooks, 1–15. https://doi.org/10.1016/b978-0-323-91268-6.00007-7
©BiologyOnline.com. Content provided and moderated by Biology Online Editors.