Nonsense mutation
n., plural: nonsense mutations
[ˈnɑnsɛns mjuˈteɪʃən]
Definition: a mutation that leads to a premature stop codon
Table of Contents
A nonsense mutation is the type of point mutation that renders the translation process useless by coding for a stop/nonsense codon. Since this would lead to the generation of an early truncated or non-functional protein, an mRNA-mediated decay process degrades the useless mRNA even before it’s translated.
Genome is a dynamic entity. Although commonly perceived as static because of the widely known and relatively stable nature of DNA, the genome is subject to an array of heritable genetic changes. Heritable genetic changes in the nucleotide sequence/s of the genome are called genetic mutations.
By the end of this article, you will not only be able to define nonsense mutation but also know where to place this type of mutation, the mechanism by which it develops, and answer the common questions like how it’s different from the other types of point mutations in the coding region of the genome, which type of mutation always produces a stop codon, and how do mutations affect an organism…
Nonsense Mutation Definition
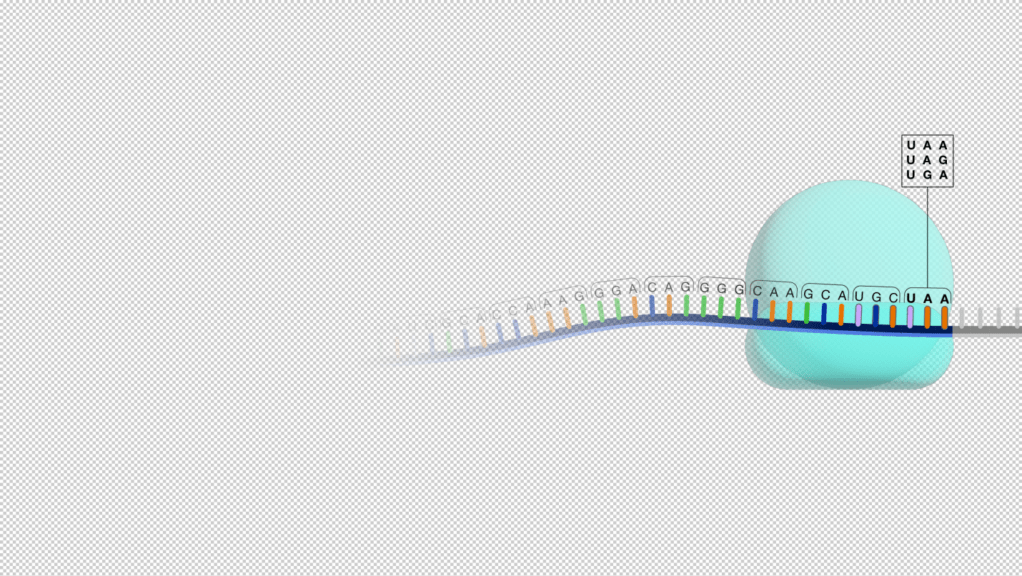
What is a nonsense mutation? In biology, a nonsense mutation is a type of point mutation arising due to replication error and resulting in the generation of a nonsense codon/stop codon. This stop codon does not code for amino acid and leads to a protein product that is early truncated (shortened protein) and the resulting protein is thereby non-functional. Since the nonsense point mutation codes for a premature stop codon, it can also be called stop mutation.
Mutations help in giving rise to alternative forms of the same gene. This process of occurrence of genetic mutation is called mutagenesis while the organism in which a mutation manifests in the form of a phenotypic change is called mutant. Now, after this brief description of the mutation process, let’s learn in detail about nonsense mutations. We will cover the different types of mutations of which one type is a nonsense mutation.
Watch this vid about nonsense mutations:
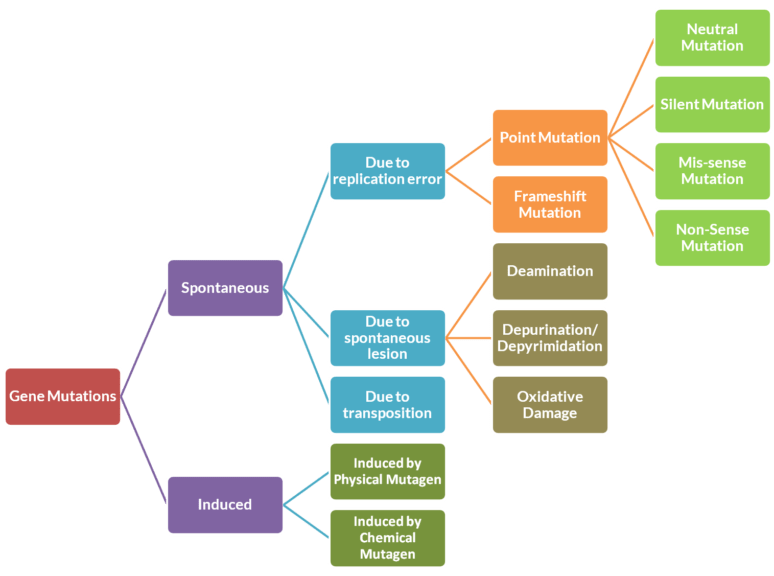
Types of Mutations and Placement of Nonsense Mutation
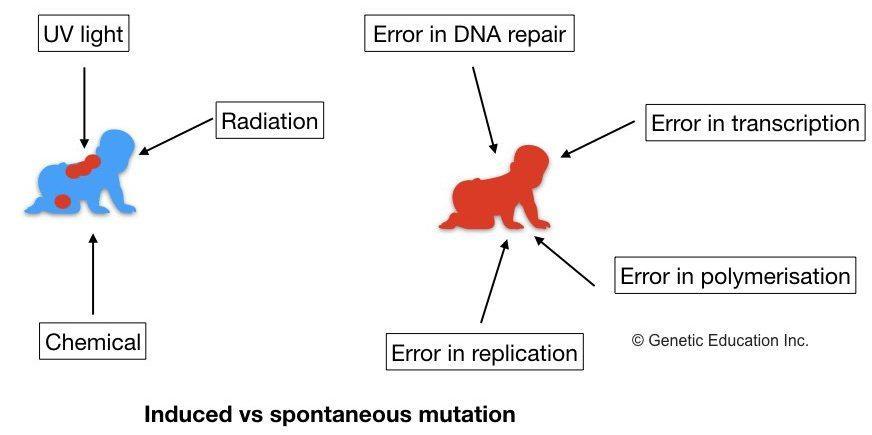
Mutations can be classified into two types — spontaneous and induced mutations. Spontaneous mutations are the ultimate source of natural genetic variations in populations. They develop spontaneously. They develop even in the absence of a mutagen (mutagenic agent). On the other hand, induced mutations don’t develop spontaneously and are induced by physical, biological, or chemical mutagens.
Note it!
A nonsense mutation is placed in the ‘spontaneous mutation’ broad category.
Mechanisms Causing Spontaneous Mutations
There are three mechanisms by which spontaneous mutations can arise. They are:
- Due to replication error – two subtypes: Point mutations and Frameshift mutations
- Due to spontaneous lesion – two subtypes: Deamination, Depurination/Depyrimidination, Oxidative damage
- Spontaneous mutations
Biology definition:
A nonsense mutation is a type of mutation resulting in a nonsense codon. The nonsense codon, as the name implies, would not code for an amino acid. A possible outcome is a protein product that is early truncated, incomplete, and often nonfunctional. Genetic disease, such as Duchenne muscular dystrophy, is caused by a nonsense mutation in a gene responsible for a specific protein, for example, dystrophin.
Compare: missense mutation, silent mutation
See also: point mutation, mutation, substitution mutation
Point Mutations Overview
Point mutations (literally stand for mutation at a definite point) are the mutations where only one nucleotide change is involved. Point mutations can occur in different parts of the genome.
- If point mutation occurs in the non-coding region of the genome, it doesn’t result in an altered amino acid sequence during the translation process.
- If point mutation occurs in the coding region of the genome, it holds the potential to alter the gene sequence/s and thereby altering the amino acid it codes for. This eventually alters the protein sequence, structure, and function. This at last results in an apparent change in gene expression.
Note it!
A nonsense mutation is placed in the ‘mutation due to replication error’ sub-category of spontaneous mutations. It is further placed in the ‘point mutation’ sub-category of mutation due to replication error (specifically, in the sub-category of point mutations – coding region).
Outcomes of a Nonsense Mutation
A nonsense mutation in a gene can result in different types of outcomes ranging from deleterious to neutral to beneficial. Let’s briefly look at the different nonsense mutations.
Deleterious
- Nonsense mutations that lead to the truncation of a protein that performs a vital function in the body will turn out to be deleterious mutations. When such nonsense mutation occurs, it leads to an overall decline in the reproductive fitness and success of the organisms, hence sometimes called harmful mutations.
- Nonsense mutations of this nature are the most common ones.
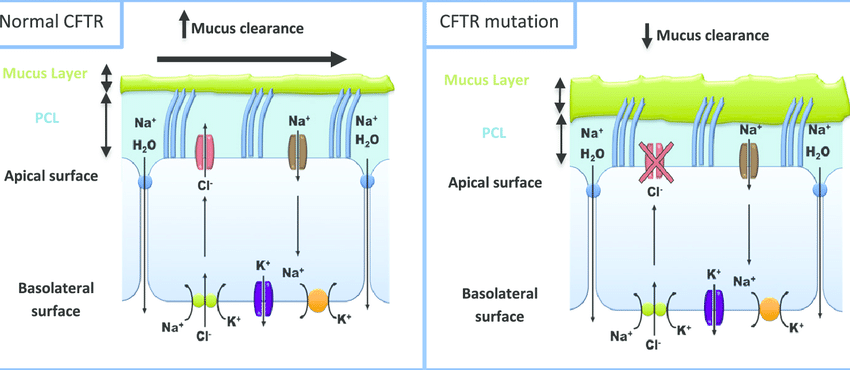
- Example: Mutation in the ion channel protein (CFTR protein) or alpha-globin and beta-globin producing genes (hemoglobin protein).
Neutral
- Nonsense mutations that go undetected due to no apparent change in the protein functioning and effects are recognized as neutral in nature. When such nonsense mutations occur, the organism is neither positively affected nor negatively affected.
- Nonsense mutations of this nature are less common than deleterious ones.
- Example: Mutation at the site exactly before the very last amino acid of a protein.
Beneficial
- Nonsense mutations that lead to an overall increase in the reproductive fitness and success of the organisms are recognized as beneficial in nature.
- Nonsense mutations of this nature are the least common ones.
- Example: Mutation at a site that affects a channel protein in selectively eliminating the transportation of toxins from external to cell’s internal environment.
So, now when asked “what is the effect of a nonsense mutation”, we can explain each of the above situations.
Nonsense Mutation Example
Let’s try to put forth a hypothetical example to make you understand the ‘nonsense mutation concept’.
- Example to understand the difference between “Nonsense mutation” and the other 3 types
Consider we have a hypothetical codon ABC coding for arginine (a basic amino acid).
- Change from ABC to ABB: ABB also coding for arginine. This is a silent mutation. This change at the genotypic level didn’t change the amino acid coded or any change in the protein that the amino acid further codes for.
- Change from ABC to AAA: AAA coding for lysine. Lysine is another basic amino acid. This is a neutral mutation. This change at the genotypic level changed the amino acid coded (from arginine to lysine). But, the nature of amino acids is the same (both are basic). This rarely changes the protein coded for or the protein function.
- Change from ABC to AAC: AAC coding for aspartate. Aspartate is an acidic amino acid. This is a missense mutation. This change at the genotypic level changed the amino acid coded (arginine to aspartate) and also changed the nature of amino acids (basic to acidic). This will change the protein coded for and the protein function.
- Change from ABC to ACC: ACC coding for stop codon/nonsense codon. This is called a nonsense mutation. This change at genotypic level codes for “NO AMINO ACID”. No amino acid means that the protein product would either be incomplete or non-functional or not formed at all. This results due to the early truncation and leads to the termination of the translation process.
Note it!
- The alteration in the nonsense mutation involves change at the DNA level. This mutated DNA is sometimes referred to as nonsense DNA too.
- The protein coded by the disrupted nonsense DNA is sometimes referred to as nonsense protein too.
- Synonymous versus Non-synonymous mutation
If asked to place nonsense mutation in one of these categories, where will you place it; synonymous or non-synonymous?
- Synonymous mutation: No change in amino acid. (Example: silent mutation)
- Nonsynonymous mutation: Change in amino acid/amino acid mutations/ different amino acid (Example: neutral mutation, missense mutation, nonsense mutation)
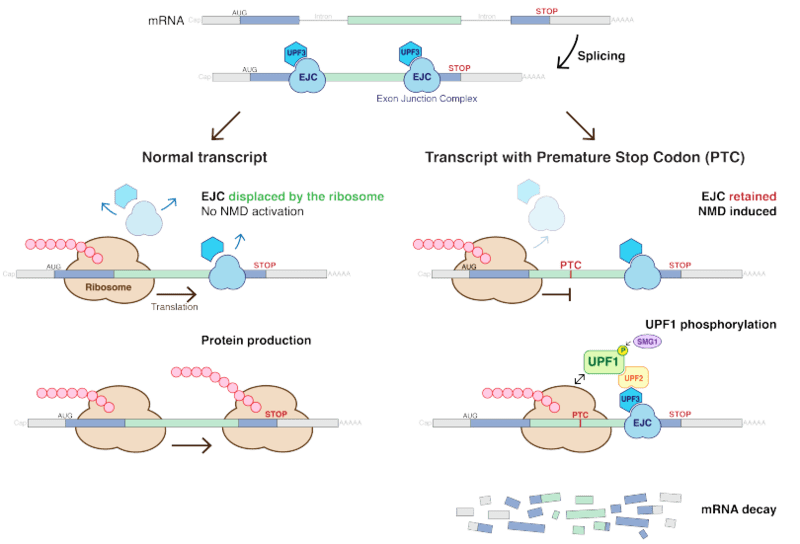
Nonsense-mediated mRNA decay
Nonsense-mediated mRNA decay is a process by which an organism’s body ensures to degrade all the premature/truncated/shortened proteins. More common than premature protein degradation is the decay of the mRNA itself that is containing these nonsense mutations.
This nonsense-mediated mRNA decay occurs in many organisms like human beings, yeasts, etc. This mechanism ensures that the nonsense variant of mRNA which can potentially lead to severe genetic diseases or genetic disorders is completely degraded before it is translated to useless non-functional proteins. It can be called a smart energy saver and surveillance step.
Pathology Associated with Nonsense Mutations
Nonsense mutations have the potential to cause a range of genetic diseases or disorders since they prevent the successful completion of the translation process of some functionally vital proteins. An important point to note here is that these diseases can be caused by some other type of mutations too in the same gene.
Some popularly known pathologies associated with nonsense mutations are:
Cystic fibrosis:
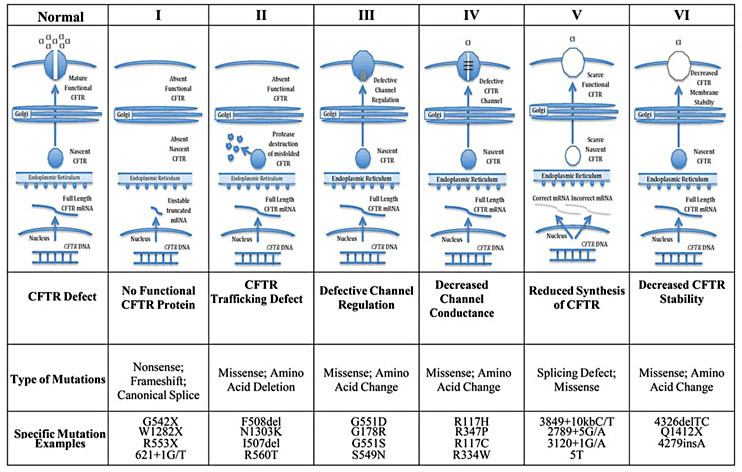
It is caused due to a nonsense mutation in the CFTR protein (an ion channel membrane protein) at position 542. The mutation is called G542X mutation as the codon coding for glycine (G) is replaced by a stop codon (X).

Figure 6: Cystic fibrosis can result from a number of mutations. One of these is a nonsense mutation called G542X as can be noticed in Class-I where no functional CFTR protein is synthesized. Image Credit: Jennings, M. T., 2014. Beta thalassemia:
It is caused due to a nonsense mutation in the beta-globin chain of the hemoglobin protein. Since hemoglobin is the main oxygen-carrying protein in the RBCs, patients with beta-thalassemia have compromised oxygen levels reaching their vitals.

Figure 7: Beta thalassemia can result from nonsense mutations in the beta-globin gene at any of the different sites of the 3 exons that constitute this gene. Sites are marked in the picture. Image Credit: Kazazian H. H., Jr (2021). Hurler syndrome:
It is caused due to a nonsense mutation in the IDUA gene that codes for the alpha-L-iduronidase enzyme. This enzyme is responsible for the breakdown of GAGs (a complex sugar). A deficit or absence of this enzyme can be detrimental to the limit of death in children.

Figure 8: Buildup of GAGs leads to Hurler syndrome due to a nonsense mutation that disrupts the production of GAG-hydrolyzing IDUA enzyme (alpha-L-iduronidase enzyme). Image Credit: Newbornscreening.info.
Duchenne muscular dystrophy:
It is caused by a nonsense mutation in the DMD gene coding for dystrophin protein. This is more common in males than females. Ataluren is currently being used to treat Duchenne muscular dystrophy.

Figure 9: Ataluren is being used to treat Duchenne muscular dystrophy. Image Credit: Haas, M., 2015.
Mastering Four Gene Mutation Types
People are often confused between these terminologies of silent, neutral, missense, and nonsense mutation. Let us put it objectively on the table for you! ☺
Based on the change/s in the functionality of the genes due to point mutations, point mutations can be classified into 4 types.
They are:
In this table, learn the four types of point mutations, silent, neutral, missense, and nonsense mutation, which all have fine line differences. Understanding these differences can bring a lot of clarity on how each one of them is stupendous in itself.
Table: The Four Types of Point Mutations | ||||
|---|---|---|---|---|
| Characteristic Feature | Silent Mutation | Neutral Mutation | Missense Mutation | Nonsense Mutation |
| Change at the genotypic level (in DNA sequence) | Yes | Yes | Yes | Yes |
| Types of genotypic change | Synonymous codon developed | Non-synonymous codon developed | Non-synonymous codon developed | Nonsense codons/ premature termination codons/ premature stop codon developed |
| Change at the amino acids level | No | Yes | Yes | Yes |
| Nature of amino acid encoded | Same | Same | Different | Stop codon/ Premature termination codon |
| Nature of protein encoded/Change in protein synthesis | Same | Same | Different | Incomplete/ Non-functional protein product |
Data Source: Dr. Harpreet Narang of Biology Online
Answer the quiz below to check what you have learned so far about nonsense mutations.
References
- Jennings, M. T., Riekert, K. A., & Boyle, M. P. (2014). Update on key emerging challenges in cystic fibrosis. Medical principles and practice : international journal of the Kuwait University, Health Science Centre, 23(5), 393–402. https://doi.org/10.1159/000357646
- Thein SL. The molecular basis of β-thalassemia. Cold Spring Harb Perspect Med. 2013 May 1;3(5):a011700. doi: 10.1101/cshperspect.a011700. PMID: 23637309; PMCID: PMC3633182.
- Kazazian H. H., Jr (2021). A Long, Fulfilling Career in Human Genetics. Annual review of genomics and human genetics, 22, 27–53. https://doi.org/10.1146/annurev-genom-111620-095614
- Haas, M., Vlcek, V., Balabanov, P., Salmonson, T., Bakchine, S., Markey, G., Weise, M., Schlosser-Weber, G., Brohmann, H., Yerro, C. P., Mendizabal, M. R., Stoyanova-Beninska, V., & Hillege, H. L. (2015). European Medicines Agency review of ataluren for the treatment of ambulant patients aged 5 years and older with Duchenne muscular dystrophy resulting from a nonsense mutation in the dystrophin gene. Neuromuscular disorders : NMD, 25(1), 5–13. https://doi.org/10.1016/j.nmd.2014.11.011
- Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E (1 August 2003). “Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways”. The EMBO Journal. 22 (15): 3960–70. doi:10.1093/emboj/cdg371. PMC 169044. PMID 12881430
©BiologyOnline.com. Content provided and moderated by Biology Online Editors.