Phosphodiester bond
n.
Definition: formed when exactly two hydroxyl groups in phosphoric acid react with a hydroxyl group on other molecules forming ester bonds
Table of Contents
Phosphodiester Bond Definition
Phosphodiester bonds are the backbone of the strands of nucleic acid present in the life existing on Earth. Phosphodiester bond is formed when exactly two hydroxyl groups in phosphoric acid react with a hydroxyl group on other molecules forming ester bonds. We can also define a phosphodiester bond as the bond which occurs when phosphate forms two ester bonds.
What is a phosphodiester bond?
The phosphodiester bonds are formed as the result of the condensation reaction between phosphate groups and hydroxyl groups of two sugar groups. For instance, the group that is being formed by the bonding of one oxygen atom and one hydrogen atom is called the hydroxyl group. Such groups are written as -OH or -HO. The “–” represents the carbon to which the hydroxyl group will be attached. Moreover, the molecules containing a single atom of phosphorus covalently bonded to four oxygen atoms are called phosphate groups. The other name for the phosphodiester bond is phosphoester bond.
A phosphodiester bond is a chemical bond that forms when exactly two hydroxyl groups in phosphoric acid react with a hydroxyl group on other molecules forming ester bonds. It is found in the DNA and RNA backbone.
Are phosphodiester bonds covalent?
Phosphodiester bonds are covalent in nature and thus are formed by the sharing of electrons between their atoms. Moreover, the enzymes that usually break the phosphodiester bonds are called phosphodiesterase abbreviated as PDE. The PDE enzyme families are classified into twelve subfamilies that are based upon the amino acid sequences, substrate specificities, regularity, and pharmacological properties, and finally the tissue distribution. (Ref. 1).
Phosphodiester Bond Formation
What is a phosphodiester bond formed between groups or molecules? Phosphodiester bonds are formed due to the reaction in between the hydroxyl groups of two sugar groups and a phosphate group and thus, oligonucleotide polymers are formed as the result of a combination of the diester bond in the phosphoric acid and the sugar molecules present in the DNA and RNA backbone.
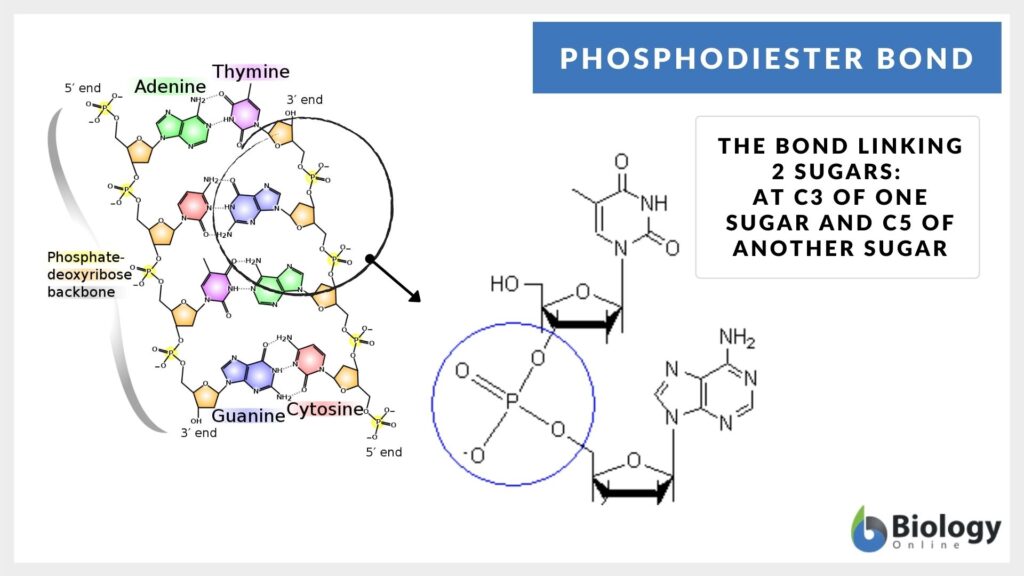
The 3′- carbon is linked with the 5′- carbon in the DNA and RNA via the phosphodiester bonds and thus they act as the backbone of nucleotides. These are the bonds that hold the sugar-phosphate components of the DNA molecule together. The schematic diagram of the formation of the phosphodiester bond is elaborated in Eqs 1.

It can be seen in Eqs 1 that during the reaction in between the -OH groups of phosphoric acid and other molecules, a couple of ester bonds are formed in the phosphodiester group. The ester bonds are formed as the result of a condensation reaction in which the water molecule is lost.
During the polymerization of the nucleotides, so that the nucleic acids are formed, the -OH in the phosphate groups gets attached to the 3′- carbon of the sugar of one nucleotide to the phosphate of the other available nucleotide, and hence, the ester bonds are formed.
The nucleotides are formed via a nitrogen base (adenine, guanine, thymine, uracil, or cytosine), a pentose sugar, and a phosphate molecule (PO4-3). Thus, after the successful elimination of the water molecule, a linkage is formed that has been referred to as the phosphodiester linkage. The structure of ribonucleoside triphosphate formation and the phosphodiester bond formation can be seen in Fig 1.

The polymerization of RNA and DNA occurs by the process of condensation of two monomers or the strands of DNA and RNA with the nucleotide triphosphate. The said condensation reaction, in nature, is very similar to the peptide condensation reactions and as a result, a single nucleic acid strand is formed. This nuclei strand is a phosphate pentose polymer also written as the polyester that has purine and pyrimidine bases as the side groups.
How do the nucleotides bond? The links between the nucleotides are the phosphodiester bonds (nucleotide phosphodiester bond). As far as the anatomy of the chemical reaction is concerned, there is a free -OH group available at the 3′ end present at the 3′- carbon of the sugar. Similarly, there is a free phosphate group present at the 5′ end present at the 5′- carbon of the sugar. Thus, the synthesis of the bond will proceed to the 3’- end via 5’- end. And so, the convention sequences are written in the 5’->3’- direction.
Watch the video below to get an overview of the structure of DNA and RNA and find out where phosphodiester bond forms.
Phosphodiester bonds of DNA and RNA
The polymeric diesters of phosphoric acids are the nucleic acids that are very capable to store and transfer biological information. Cleavages of the phosphodiester linkages by the enzymes of protein and nucleases are a very important biological process and hence, catalytic effectiveness of the nucleases and the ribonucleic acids has made the phosphodiester cleavages to be center of research in many internationally recognized research laboratories. (Ref. 2)
What bond is cleaved by during the first reaction of integration? In the biological systems, the diester linkage of 5′-O of one nucleotide to the 3′-O of another are cleaved by many of the diversified enzymes. Thus, depending upon the types of enzyme used, the hydrolyzation of the phosphodiester bond of DNA occurs, either to the 5′- phosphate group or 3′- whereas contrary to it, the RNA bonds, with minor exceptions, undergo the cycle of transesterification to a 2’- or 3’- cyclic phosphate that ultimately gets hydrolyzed to 2’- or 3’- phosphate groups. The schematic transformation that occurs during this process of phosphodiester linkages has been elaborated in Fig 2.
It has been confirmed by various research studies that under the physiological conditions, the 3’- and 5’- phosphodiester linkages are extremely stable even without the employment of the catalysts. Furthermore, it has been estimated that the half-life of the hydrolysis of a phosphodiester bond in DNA at room temperature is 30 million years, which elaborates that the phosphodiester cleavage is accelerated by the protein enzymes and nucleases by a factor of 10016 (Ref. 3).
Similarly, the phosphodiester linkages available in the RNA molecule are extremely labile due to the presence of hydroxyl functions that serve as the intramolecular nucleophile thus leading to the transphosphorylation by the departure of 5’-linked nucleoside. Additionally, its half-life at room temperature and pH of six to seven is approximately 120 months and the enzymatic cleavage is also quicker.
It is a very interesting fact that the RNA phosphodiester bonds are endangered to the cleavage by RNA itself by the arrangements known as ribozymes. Hence, the length of these sequences ranges from seventy to hundreds of small ribozymes to the chains of extensively large ribozymes. Thus, the catalytic activities of such sequences are somewhat modest to that of the enzymes of proteins. (Ref. 4).

Mechanism of Phosphodiester Formations
A firm understanding of the mechanism behind the hydrolysis and transesterification reactions of the nucleic acids can be achieved by going through the phosphodiester model studies. Such information is very vital for the in-depth evaluations of the mechanism of extremely complicated enzymatic reactions that are achieved via artificial cleaving agents, that although have catalytic activities like enzyme but are not robust.
The most important parameters that are studied to recognize the mechanisms behind the formation of phosphodiester bonds include the profiles of pH rates, relationships of linear free energies, and the experimental approaches of the kinetic heavy and dissection of data of detected rate constants toward the formation of various ions. Thus, the effects of deprotonation and protonation of a particular atom are obtained. Similarly, the position of the transition state on the reaction coordinate is determined via linear free energy relationships. Hence, the information on the charge distribution in the transition state can be obtained by analyzing the polar properties of the entering or departing nucleophile or sometimes the non-departing groups, and then the reaction rate effects are compared with the effects of equilibrium of the undergoing reaction. Therefore, it can also be predicted whether the transition state is early which means it is close to the reactants, or late that states that it is very near to the products. Lastly, the free energy relationship is obtained as a result of generating a plot in between the activation free energy against the change in standard free energy of the reaction.
The two most common mechanisms that are used to elaborate on the formation of phosphodiester bonds are kinetic heavy atom isotope effect (KIE) and kinetic solvent isotope effect (KSIE). The KIE model can be used for both enzymatic and non-enzymatic reactions whereas the KSIE is employed to distinguish between the alternative mechanism. (Ref. 5)
How many phosphodiester bonds are in DNA?
It is very important to know the methodology to calculate the number of phosphodiester bonds available in various DNA molecules. The number of phosphodiester bonds present in the DNA is proportional to the number of base pairs or nucleotides present in it. Hence, after thorough research works, scientists have presented a formula to calculate the number of phosphodiester bonds (PD). The formula is as under:
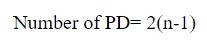
where n is the no. of base pairs or nucleotides.
To further understand the formula let us calculate the number of PD bonds present in the 5’-CTAGAG-3’. Here the number of base pairs is six hence the value in n is 6.
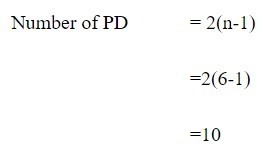
Thus, in 5’-CTAGAG-3’ there are ten available phosphodiester bonds. Similarly, in 3’-GATCT-5’ the number of phosphodiester bonds present in the DNA is 8.
Phosphodiester Bond – Biological Importance
The impact of phosphodiester bonds in the life of living organisms can never be overlooked. They have always proved to be the center of life on earth. Moreover, they act as the phosphodiester backbones of the strands of the nucleic acids. Thus, they are very vital in carrying out the information of the cell and after directing the process of synthesis of proteins, the inherited characteristics of the living things are determined.
Deoxyribonucleic acids (DNA) and ribonucleic acid (RNA) are the two main classes of nucleic acids. DNA acts as the blueprint of life in all free-living organisms while RNA is the genetic material of the majority of the viruses while it also plays a very significant role in many processes in the human bodies such as the making of proteins. Thus, it can be concluded that the swift formation of phosphodiester bonds is very vital in maintaining the structures of the nucleic acids that further plays a momentous role in the formation of DNA and RNA as for the replication of DNA and synthesis of RNA it is very vital to have a directionality of the nucleic acids.
Moreover, Lesch Nyhan syndrome, mitochondrial depletion syndromes, and ataxia-telangiectasia are some of the diseases that are caused by the defects in the nucleotide salvage enzymes. Although, there are some treatment options available to address the symptoms of such diseases unfortunately there is no cure. (Ref. 6)
Summary
It can be concluded from the above thorough discussion that the linkage of phosphoric acid molecules (H3PO4) with the two hydroxyl groups (-OH or -HO) gives rise to the formation of phosphodiester bonds and linkages. Moreover, it has been confirmed that the nature of such bonds is covalent. The two very important chemical reactions that play a very vital role in its formation are the condensation reaction and the polymerization reaction. Moreover, KIE and KSIE are the two very imperative methods that are used to understand the formation of phosphodiester bonds. The phosphodiester bonds are found in the nucleic acids; thus, their main recompenses are to control their functionality, structure and formations. The polymeric diesters of phosphoric acids are the nucleic acids that are very capable to store and transfer the biological information. Furthermore, cleavages of the phosphodiester linkages by the enzymes of protein and nucleases is a very important biological process and thus has been the center of research for many years. The anatomy of the chemical reaction through which the phosphodiester bonds are formed — there is a free -OH group available at the 3’ end present at the 3’- carbon of the sugar. Similarly, there is a free phosphate group present at the 5’ end present at the 5’- carbon of the sugar. The number of the bonds present in the DNA is proportional to the number of base pairs or nucleotides present in it and can be calculated by the formula as number of PD= 2(n-1). The impact of phosphodiester bonds in the life of living organisms can never be overlooked. They have always proved to be the center of life on earth. Moreover, they act as the phosphodiester backbones of the strands of the nucleic acids.
Try to answer the quiz below to check what you have learned so far about the phosphodiester bond.
References
1. Phosphodiester bond fromation. (2016). Biosyn.Com. https://www.biosyn.com/tew/What-is-a-Phosphodiester-bond.aspx#:~:text=A%20phospodiester%20bond%20is%20a,groups%20and%20a%20phosphate%20group.&text=The%20phosphodiester%20bond%20links%20a,carbon%20in%20DNA%20and%20RNA.
2. Mikkola, S., Kaukinen, U., & Lönnberg, H. (2001). The Effect of Secondary Structure on Cleavage of the Phosphodiester Bonds of RNA. Cell Biochemistry and Biophysics, 34(1), 95–119. https://doi.org/10.1385/cbb:34:1:95
3. Linjalahti, H., & Mikkola, S. (2007). Intra- and Intermolecular Interactions Influence the Reactivity of RNA Oligonucleotides. Chemistry & Biodiversity, 4(12), 2938–2947. https://doi.org/10.1002/cbdv.200790243
4. Mikkola, S., Lönnberg, T., & Lönnberg, H. (2018). Phosphodiester models for cleavage of nucleic acids. Beilstein Journal of Organic Chemistry, 14, 803–837. https://doi.org/10.3762/bjoc.14.68
5. Kinetic Isotope Effects Quantify pH-Sensitive Water Dynamics at the Pt Electrode Interface. (2020). The Journal of Physical Chemistry Letters. https://pubs.acs.org/doi/10.1021/acs.jpclett.0c00185
6. Fasullo, M., & Endres, L. (2015). Nucleotide Salvage Deficiencies, DNA Damage and Neurodegeneration. International Journal of Molecular Sciences, 16(12), 9431–9449. https://doi.org/10.3390/ijms16059431
©BiologyOnline. Content provided and moderated by BiologyOnline Editors.