Phylogeny
n., [faɪˈlɑdʒəni]
Definition: Evolutionary history of a group of organisms
Table of Contents
Phylogeny refers to the evolutionary history of the development of a species or of a taxonomic group of organisms. The phylogenetic relationships are depicted in the form of a phylogenetic tree, i.e. a tree diagram depicting how one taxon is closely or distantly related to another taxon. The tree diagram demonstrating phylogenetic relationships is based on molecular sequencing data analyses as well as on morphological data matrices.
Phylogeny Definition
The definition of phylogeny in biology pertains to the evolutionary history or development of a group of organisms, such as a tribe or a racial group. The phylogeny meaning is somewhat similar to the term phylogenesis and as such, they are sometimes used interchangeably. However, in a stricter sense, phylogenesis refers to the biological process by which a particular taxon exists. Phylogenetics is another related term.
It refers to the scientific study of phylogeny. It applies molecular and analytical methods in understanding evolutionary history and processes involved during the development of a species or of a taxon. Phylogeny is the result of the studies and the analyses of evolutionary occurrences of living organisms and it is represented by a tree diagram called the phylogenetic tree. Phylogenetics attempts to explain the evolutionary relatedness among various groups of organisms through molecular sequencing data and morphological data matrices.
Etymology
The term phylogeny was derived from German Phylogenie that was coined by Ernst Haeckel in 1866. Phylogenie, in turn, came from the Greek words φῦλον (phûlon), meaning “tribe”, “genus”, or “species” and -γένεια (-géneia, -geny), meaning “generation” or “production”. Synonyms: phylogenesis.
Phylogeny vs. Ontogeny
Both phylogeny and ontogeny deal with the origin and the development of organisms. They are both concerned with developmental histories. However, ontogeny is different from phylogeny in a way that it looks through the historical development of an organism within its own timeline (e.g. from its simplest to the most complex form) and not on its evolutionary history. Thus, ontogeny is to the development of an individual organism as phylogeny is to the evolution of a species.
Phylogeny vs. Taxonomy
While phylogeny is concerned with the evolutionary relatedness and history of organisms, it is not concerned with the identification of these organisms. Rather, it is the main concern of taxonomy. To be more precise, taxonomy is the branch of science that is concerned chiefly with identifying, naming, and classifying organisms. It puts organisms into taxonomic ranks, e.g. domain, kingdom, phylum or division, class, genus, species. Nevertheless, the classification is based on the morphology and phylogeny of organisms, and phylogenetics provides information for use during the identification and classification of organisms. So while taxonomy is concerned chiefly with the identification and classification of organisms, phylogeny provides data for such purpose and a phylogenetic classification would be one that is largely based on molecular phylogeny data.
Molecular Phylogeny
Molecular phylogeny, a branch of phylogeny, makes use of molecular sequencing to study evolutionary relationships and histories. Molecular sequencing, in this regard, is a useful tool to understand the phylogenies of different taxa. Basing the relatedness on morphology, anatomy, physiology, and life cycles can be confounding. There are instances wherein some traits are easily identified as either similar or disparate. However, there are also instances when their similarities and disparities are vague. Furthermore, there are also situations when two species seem to belong to a common taxon but after analyzing their genomes they turned out to be evolutionary distant.
Fortunately, more advanced tools for study and research have become available and they have provided scientists a more reliable basis for determining and analyzing phylogeny. Nucleic acids, such as DNA and RNA, store and retain certain genetic information that scientists use as hints of plausible evolutionary origins and history. It is because these biomolecules are heritable.
By comparing such information through the aid of a computer program, the degree of relatedness between and among organisms can be recognized. By looking at the genome (as well as the proteins being coded for), the evolutionary relatedness can be analyzed, i.e. whether the organisms are closely related or distant. One of the most widely used for molecular phylogeny studies and analyses is the sequence of the small subunit of the ribosomal RNA. (1) The analyses that can be derived from such sources have the advantage of providing quantifiable data. The relatedness between taxa can be demonstrated through molecular sequencing data and morphological data matrices.
Phylogeny Diagrams
Phylogeny may be represented by a tree diagram called phylogenetic tree (also called an evolutionary tree). The diagram depicts the relationships among organisms or the relatedness between taxa. It is created based on molecular phylogeny studies and on morphological data. By comparing organisms based on commonalities and incongruences, their evolutionary relationship could be established and represented in a tree diagram.
A phylogenetic tree may be rooted or unrooted. A rooted phylogenetic tree implicates a common ancestor where closely related taxa descended from. An unrooted phylogenetic tree, in contrast, does not show a common ancestor but it hypothesizes on the degree of evolutionary relatedness between taxa. The tree diagram is essential as it aids in understanding biodiversity, evolutions, genetics, and ecology of the various groups of organisms. By simply looking at the positioning and the length of the ‘branches’, one could easily infer how one group may be evolutionarily related to another. Those that are joined together implicate evolutionary relatedness. The internal nodes signify a hypothetical common ancestor.
Tree of Life
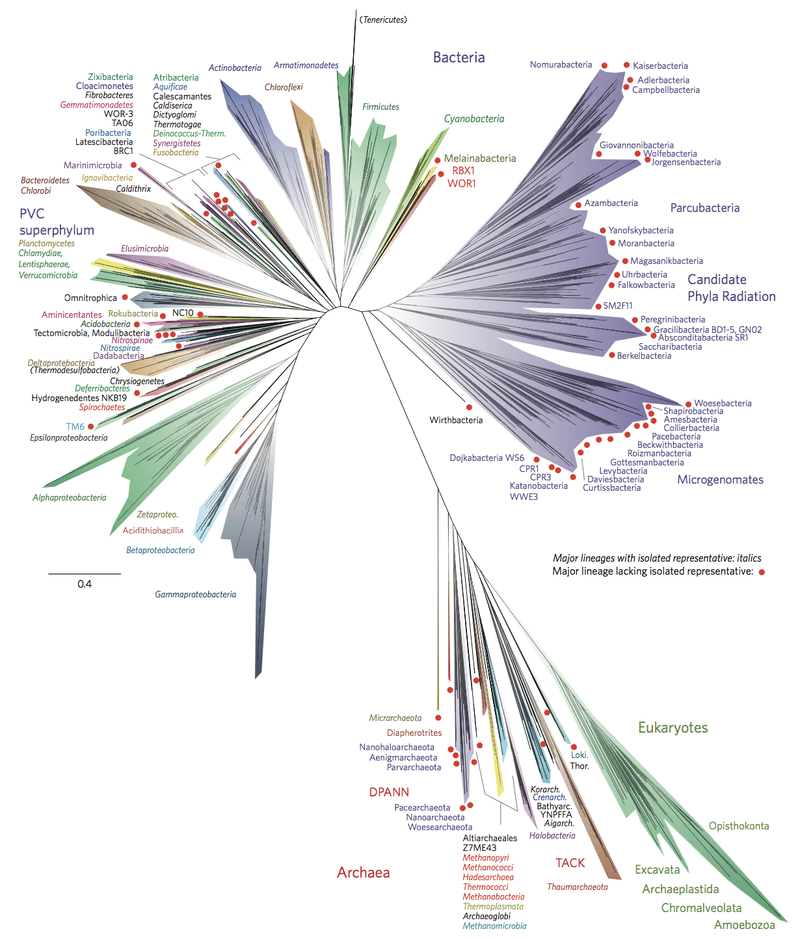
In biology, the Tree of life is a schematic model that shows the evolution of organisms both the extinct and the living. Tracing the bottom (the root of the tree) leads to the last universal common ancestor of life on earth. As for the tips, they represent the living organisms at present and some being the most recent in the evolutionary lineage. In 2016, a modern metagenomics tree of life was suggested. (2) The diagram includes 92 bacterial phyla, 26 archaeal phyla, and all five of the Eukaryotic supergroups.
Microbial Phylogeny
Microbial phylogeny is the evolutionary history or development of microorganisms, such as bacteria. Similar to any other early phylogenies, the tree diagrams used to depict evolutionary relationships were based on morphologies, and in this case, bacterial structure. In the 1960s to 1970s, microbial phylogenetics emerged and scientists began creating phylogenetic trees based on nucleic acids and protein sequencing rather than on anatomy and physiology. (3) One of the most notable contributors to microbial phylogenetics is Carl Woese. He studied the small subunit rRNA oligonucleotides of bacteria and compared them to determine evolutionary relatedness. He and his team were the first to suggest that archaebacteria were different from bacteria. (4) This led to the three-domain classification system of organisms: domain Bacteria, domain Archaea, and domain Eucarya, thereby, refuting the old prokaryote-eukaryote dichotomy. Modern phylogenetic studies reveal that there are 92 bacterial phyla. Nonetheless, this figure is not official just yet. Moreover, there are also no officially accepted taxa above the class rank in microbial classification.
Animal Phylogeny
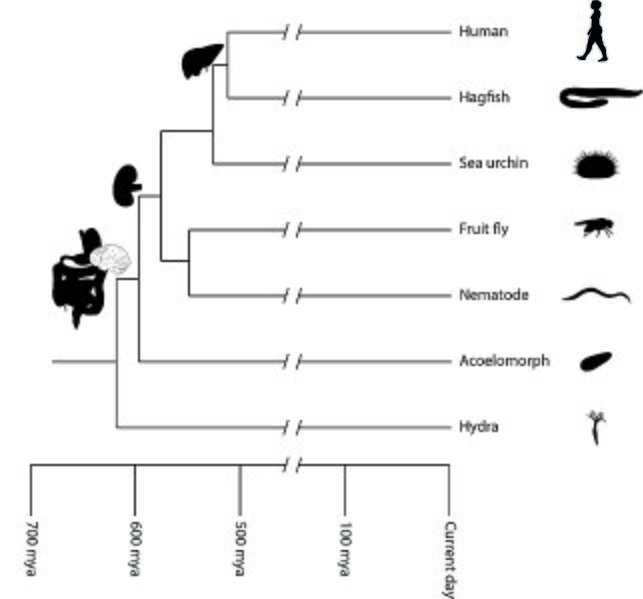
The phylogenetic tree of animals depicting the evolution of animal organs is a special phylogeny example. It shows animal phylogeny in terms of the evolution of animal organs. In this type of diagram, the evolutionary relationship of major animal lineages can be inferred based on the organ level of the organization. For instance, the digestive system seemed to first appear about 600 million years ago, and about a hundred million years later, the liver evolved in vertebrates, such as humans.
Importance
Phylogeny pertains to the evolutionary history of a taxonomic group of organisms and it is used as a basis in phylogenetics as the latter deals with the relationships of an organism to other organisms according to evolutionary similarities and differences. Thus, phylogeny is essential in the scientific study of the identification, classification, ecology, and evolutionary histories of organisms. It shows the relationships between groups of organisms (taxa), particularly the differences and similarities among them. It becomes vital in understanding biodiversity, genetics, evolutions, and ecology among groups of organisms. Apart from phylogenetics, it is also vital in the field of taxonomy. It expands the basis of evolutionary relationships of organisms from the morphological aspect to the genetic constructs of organisms.
Limitations
Inferences derived from phylogenetic studies are not absolute. The data derived from genomic studies may possibly contain erroneous data. For instance, the genomic analyses may be based on faulty data, e.g. those flawed by horizontal gene transfer between species. (5) Phylogeny that is based on several genes or proteins from different genomic sources (e.g. nuclear or mitochondrial) is also likely to be more precise than on a single gene or protein alone. Otherwise, the analysis may be a phylogeny of the gene and not of the species. Another important limitation is the lack or insufficiency of a quality DNA sample from extinct species.
NOTE IT!
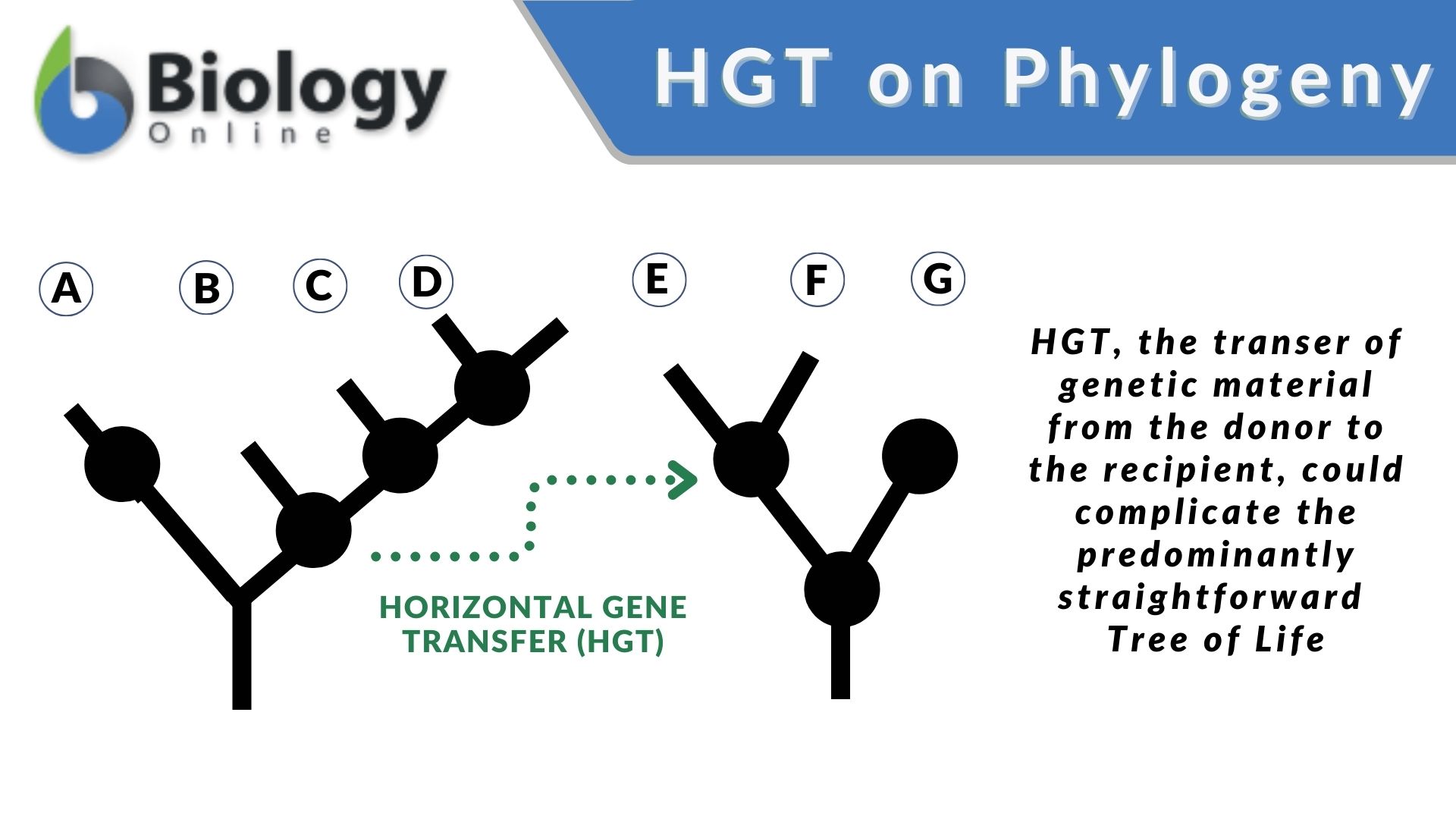
Horizontal Gene Transfer: Challenging the Tree of Life in Phylogeny
Research on horizontal gene transfer (HGT) reveals its significant and complex role in evolution and phylogeny. With the conception of advanced sequencing techniques, scientists have recognized its evolutionary and phylogenetic implications.
HGT is a phenomenon that occurs naturally, especially among bacteria and archaea. Prokaryotes are able to acquire new genetic material from other organisms, and thus, exhibit traits that make them more genetically diverse, ecologically adaptable, and competitive (e.g., antibiotic resistance, novel metabolic capabilities, ability to exploit new resources).
HGT is a way of sharing such beneficial traits. Thus, it is not surprising that newer evidence indicates that HGT may not be a rare phenomenon as previously assumed but is widespread and seemingly fundamental, especially in shaping genomes.
Not only that. HGT can cross Kingdom boundaries.
HGT appears to occur between distantly related organisms, too, like between bacteria and plants. This cross-Kingdom horizontal gene transfer between unrelated species challenges the concept of the phylogenetic, hierarchical tree of life.
Evolution is largely based on a strictly tree-like model where the evolution proceeds in a tree-like fashion with a single common ancestor for each group of organisms. Conversely, HGT suggests that genes could flow between branches of the phylogenetic tree, complicating the predominantly straightforward ancestral lines.
HGT introduces a twist.
It features the evolution of life as a complex network of interrelatedness rather than a forthright tree-like lineage. This has sparked debates in the scientific community oppugning its role in the phylogeny of organisms and up to what extent.
It’s very likely that in time it will call for a re-evaluation of how we define the species concept and shift our eyes on how we see and interpret phylogeny and the evolution of life.
Try to answer the quiz below to check what you have learned so far about phylogeny.
See also
References
- Molecular Phylogeny. (2019). Retrieved from Tulane.edu website: https://www.tulane.edu/~wiser/protozoology/notes/tree.html
- Hug, L. A., Baker, B. J., Anantharaman, K., Brown, C. T., Probst, A. J., Castelle, C. J., Butterfield, C. N., Hernsdorf, A. W., Amano, Y., Ise, K., Suzuki, Y., Dudek, N., Relman, D. A., Finstad, K. M., Amundson, R., Thomas, B. C., & Banfield, J. F. (11 April 2016). “A new view of the tree of life”. Nature Microbiology. 1 (5): 16048. DOI:10.1038/nmicrobiol.2016.48. PMID 27572647.
- Dietrich, M. (1998). “Paradox and persuasion: Negotiating the place of molecular evolution within evolutionary biology”. Journal of the History of Biology. 31: 85–111.
- Woese, C.R. & Fox, G.E. (1977). “Phylogenetic structure of the procaryote domain: The primary kingdoms”. Proceedings of the National Academy of Sciences. 75: 5088–5090.
- Woese C (2002). “On the evolution of cells”. Proc Natl Acad Sci USA. 99 (13): 8742–7.
© Biology Online. Content provided and moderated by Biology Online Editors