Gene Regulation in Eukaryotes
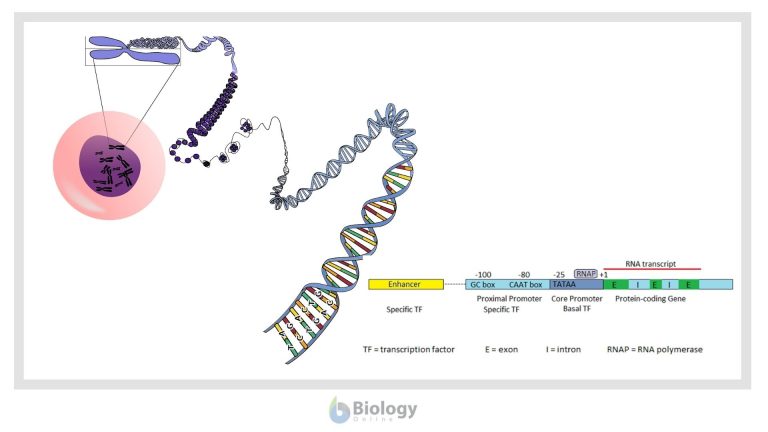
Genetic regulation in eukaryotes
Table of Contents
Reviewed by: Mary Anne Clark, Ph.D.
Eukaryotic Gene Structure
In prokaryotes the DNA is located in the cytoplasm, so that translation may begin even before transcription has been completed. Because of that, most gene regulation occurs at the level of transcription in prokaryotes. However, in eukaryotes, the DNA is separated from the cytoplasm by the nuclear envelope, so the mRNA has to find its way to the ribosomes from the nucleus. Gene activity can, therefore, be regulated at either transcription or translation, or even post-translation by modification of the translated protein. Another difference is that eukaryotic genes are not clustered into operons controlled from a single promoter. Most genes encode only a single protein, and each gene has its own promoter.
The diagram below represents the general structure of a eukaryotic gene and its major regulatory regions.
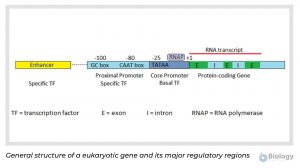
In most protein coding genes, the coding sequence is not continuous, but broken up into several exons separated by non-coding introns. The introns are removed after transcription to produce a translatable mRNA. Eukaryotic promoters include a core promoter that binds RNA polymerase and additional adjacent regions that bind other regulators. Eukaryotic promoters are quite variable and may or may not contain any or all of the three DNA binding regions represented in the image: the TATAA box in the core promoter and the CAAT and GC boxes of the proximal promoter.
Transcription Factors
Two types of protein transcription factors regulate transcription in eukaryotes. RNA polymerase binds to the core promoter with the assistance of basal transcription factors, one of which is a TATA-binding protein that recognizes the TATAA sequence of the core promoter DNA. Adjacent to the core promoter are additional regions that bind specific transcription factors. Specific transcription factors bound to the enhancer region interact with those at the promoter to enhance or inhibit transcription. Transcription factors bound to the enhancer region are active only when they are bound to the enhancer DNA. Because the enhancer region may be hundreds or even thousands of nucleotides upstream of the promoter that they regulate, the DNA must bend to allow the enhancer bound-proteins to interact with the promoter-bound proteins.
In eukaryotes another layer of regulation is necessary because of the structure of eukaryotic chromosomes. While prokaryotic chromosomes are composed of single, usually circular, molecules of DNA, in eukaryotic chromosomes the DNA is combined with proteins to form chromatin. One type of chromosomal protein is the histones, which form protein complexes around which DNA is wound to form structures called nucleosomes. These nucleosome complexes are progressively condensed with other proteins so that most eukaryotic genes are not easily accessible for transcription. The first level of eukaryotic gene regulation is the opening up of the chromosome so that both various transcription factors and RNA polymerase can get to the DNA.
MyoD and Muscle Cell Differentiation
MyoD is a transcription factor that can induce the differentiation of muscle tissue. It is only one of many regulators involved in the muscle differentiation, but will serve to illustrate the complexity of eukaryotic gene regulation. Skeletal muscle is a multinucleate tissue formed by the fusion of embryonic myoblasts. Four important regulators of muscle cell differentiation are MyoD, Myf5, myogenin and MRF4. Mice missing either MyoD or Myf5 develop normal muscle tissue, but mice that are missing both MyoD and Myf5 fail to develop any skeletal muscle at all, so these two genes work together in regulating muscle differentiation. However, mice missing the other two genes also show various muscle deficiencies so all four are important.
MyoD binds to the regulatory regions of many genes required for making skeletal muscle. It usually doesn’t act alone but forms heterodimers with similar proteins. These dimers recognize pairs of a DNA regulatory element called an E-box, whose consensus sequence is CANNTG. MyoD may play another role in initiating muscle cell differentiation because it can also remodel the chromosomes around the muscle-specific genes to make those genes accessible to transcription by binding to an enzyme that acetylates histones. Since histones are basic proteins, they are attracted to the negatively charged phosphate groups of the DNA backbone. Acetylating the histones reduces their attraction to the DNA so that the DNA can be freed for transcription.
This is an oversimplification of the roles of MyoD in activating muscle-specific genes, and MyoD itself has multiple regulators. However, it is a little window into the complexities of eukaryotic transcriptional regulation.
Post-transcriptional Regulation
Once a eukaryotic gene is transcribed, it has to be processed by removal of the introns. In larger genes with many exons, several different versions of the protein coding sequence may be produced by alternative splicing, the selection of different sets of exons to form the mRNA. In addition, the mRNA is protected from degradation in the cytoplasm by the addition of nucleotides at both the 5′ and 3′ ends of the mRNA. At the 5′ end is a methyl-guanosine cap, which helps identify the mRNA to the ribosome. At the 3′ end is a string of adenine nucleotides, the poly-A tail, which helps to stabilize the mRNA as it moves to the ribosome.
The translation of the mRNA may then be either enhanced or repressed by proteins that bind to the untranslated 5′ and 3′ regions of the mRNA. These proteins may be themselves modified in response to various factors in the cell’s environment. In addition to these regulatory proteins, regulatory RNAs may bind to a mRNA to control its translation.
After translation, the protein may be further modified. In many cases, the initiating methionine, the amino acid encoded by the start codon AUG, is removed. Some proteins, like trypsin and other digestive enzymes, are synthesized as an inactive precursor protein that has to be activated by removing part of the protein. The protein precursor of insulin is a larger protein cut into several pieces, two of which are then linked together as the A and B chains of active insulin. The alpha and beta chains of the protein hemoglobin are the products of two different genes.
Generally, transcriptional regulators select which genes are transcribed in a given cell, while post-transcriptional factors come into play to determine whether or not the encoded protein product is made.
The next lesson is on hormone production.
You will also like...

Evolution of Life – Ancient Earth
Autotrophs flourished, absorbing carbon and light. Soon after, primitive life forms that could assimilate oxygen thrived..

Sigmund Freud and Carl Gustav Jung
In this tutorial, the works of Carl Gustav Jung and Sigmund Freud are described. Both of them actively pursued the way h..
Digestion and Absorption of Food
The gastrointestinal system breaks down particles of ingested food into molecular forms by enzymes through digestion and..
Freshwater Community Energy Relationships – Producers & Consumers
This tutorial looks at the relationship between organisms. It also explores how energy is passed on in the food chain an..

Pollution in Freshwater Ecosystems
There are many environmental factors that arise due to the usage of water in one way or another and for every action tha..

Chemical Composition of the Body
The body is comprised of different elements with hydrogen, oxygen, carbon, and nitrogen as the major four. This tutorial..
